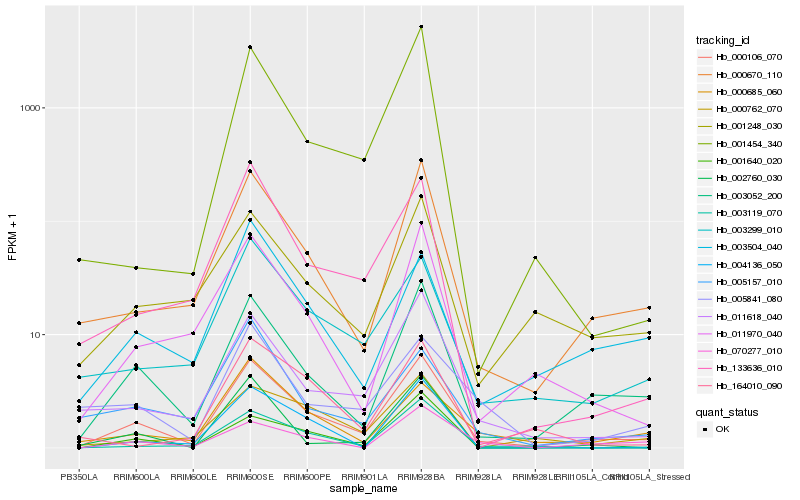
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000106_070 |
0.0 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 2 |
Hb_003052_200 |
0.115682561 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 3 |
Hb_000670_110 |
0.1364925329 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 4 |
Hb_011618_040 |
0.1604633302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003299_010 |
0.1725825451 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 6 |
Hb_011970_040 |
0.1736429084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003119_070 |
0.1753356929 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_001248_030 |
0.1871122546 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_133636_010 |
0.1897266589 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 10 |
Hb_000685_060 |
0.1972694976 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 11 |
Hb_003504_040 |
0.1984137982 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 12 |
Hb_001454_340 |
0.1993561185 |
- |
- |
ATDI21 [Manihot esculenta] |
| 13 |
Hb_002760_030 |
0.2015672214 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 14 |
Hb_070277_010 |
0.2039272303 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 15 |
Hb_000762_070 |
0.206756644 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 16 |
Hb_005841_080 |
0.207535435 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |
| 17 |
Hb_001640_020 |
0.207658183 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 18 |
Hb_005157_010 |
0.2113079635 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 19 |
Hb_004136_050 |
0.2115892286 |
- |
- |
- |
| 20 |
Hb_164010_090 |
0.2170847727 |
- |
- |
cellulose synthase, putative [Ricinus communis] |