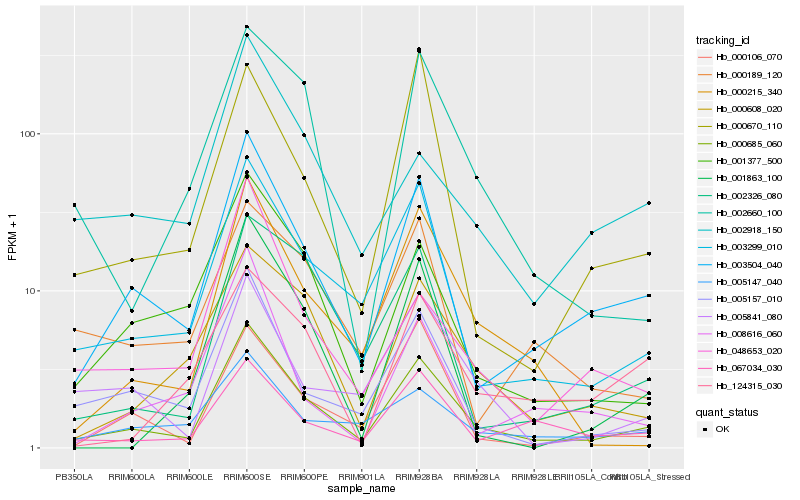
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000685_060 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_003504_040 |
0.1171595893 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 3 |
Hb_003299_010 |
0.1403839406 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 4 |
Hb_005157_010 |
0.1628576799 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 5 |
Hb_001377_500 |
0.1732171486 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 6 |
Hb_000608_020 |
0.1765352749 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 7 |
Hb_000215_340 |
0.1781617626 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 8 |
Hb_067034_030 |
0.1834846818 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_005841_080 |
0.1886515091 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |
| 10 |
Hb_002660_100 |
0.1898564485 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |
| 11 |
Hb_005147_040 |
0.1906635811 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 12 |
Hb_002326_080 |
0.1930964554 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 13 |
Hb_000106_070 |
0.1972694976 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 14 |
Hb_002918_150 |
0.2041637701 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 15 |
Hb_048653_020 |
0.20455417 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_001863_100 |
0.2047268413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000670_110 |
0.2056906166 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 18 |
Hb_124315_030 |
0.2095478446 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 19 |
Hb_008616_060 |
0.2095973492 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 20 |
Hb_000189_120 |
0.2113887907 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |