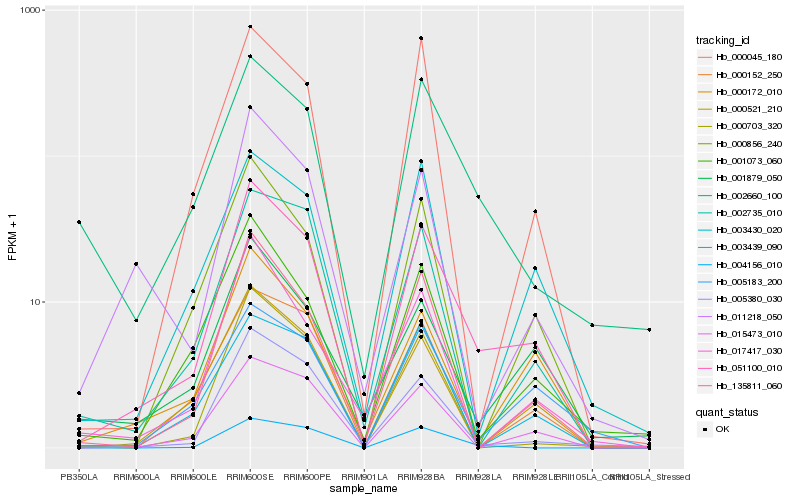
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_051100_010 |
0.0 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_135811_060 |
0.1437855242 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 3 |
Hb_000521_210 |
0.1491346092 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 4 |
Hb_005380_030 |
0.1520582691 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase B120 isoform X2 [Jatropha curcas] |
| 5 |
Hb_015473_010 |
0.1522948962 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 6 |
Hb_000856_240 |
0.1537514488 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000172_010 |
0.1564166002 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 8 |
Hb_001073_060 |
0.1646064695 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 9 |
Hb_000045_180 |
0.164775428 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 10 |
Hb_004156_010 |
0.1653751232 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 11 |
Hb_002735_010 |
0.1671336368 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 44 [Prunus mume] |
| 12 |
Hb_000152_250 |
0.1672031068 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 13 |
Hb_011218_050 |
0.1686991779 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_005183_200 |
0.1688378386 |
- |
- |
EG964 [Manihot glaziovii] |
| 15 |
Hb_000703_320 |
0.1690273678 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_001879_050 |
0.1725060754 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 17 |
Hb_017417_030 |
0.1732372811 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 18 |
Hb_003439_090 |
0.1739802434 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_002660_100 |
0.1742210517 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |
| 20 |
Hb_003430_020 |
0.175242104 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |