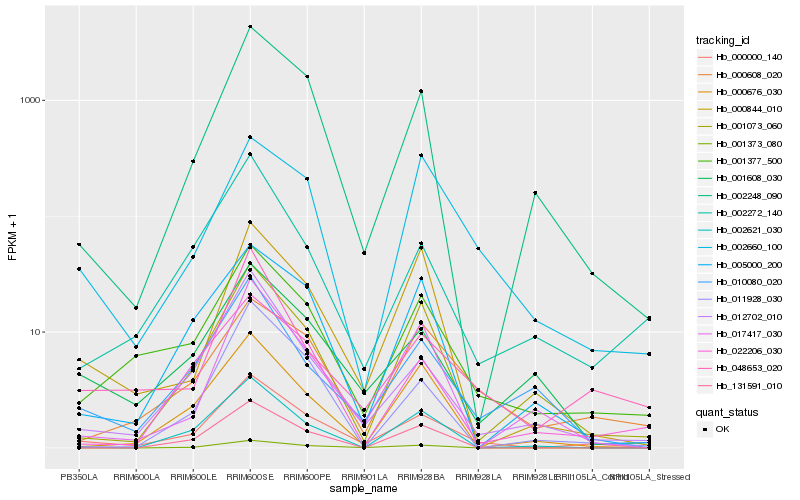
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_001377_500 |
0.1541047722 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 3 |
Hb_012702_010 |
0.168966865 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 4 |
Hb_002248_090 |
0.1800344381 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 5 |
Hb_048653_020 |
0.1878323722 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001373_080 |
0.1901884302 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001608_030 |
0.1945165649 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 8 |
Hb_010080_020 |
0.1957797337 |
- |
- |
PREDICTED: uncharacterized protein LOC105635249 [Jatropha curcas] |
| 9 |
Hb_002660_100 |
0.1974936021 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |
| 10 |
Hb_000608_020 |
0.1980769899 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 11 |
Hb_005000_200 |
0.1981173298 |
- |
- |
PREDICTED: uncharacterized protein LOC105637887 [Jatropha curcas] |
| 12 |
Hb_000844_010 |
0.2006216507 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 13 |
Hb_002272_140 |
0.2033729724 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 14 |
Hb_001073_060 |
0.2041555168 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 15 |
Hb_022206_030 |
0.2065833784 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 16 |
Hb_000676_030 |
0.2067496167 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 17 |
Hb_017417_030 |
0.2077557911 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 18 |
Hb_131591_010 |
0.2081471621 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 19 |
Hb_011928_030 |
0.209026993 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 20 |
Hb_002621_030 |
0.210658604 |
- |
- |
hypothetical protein CISIN_1g006155mg [Citrus sinensis] |