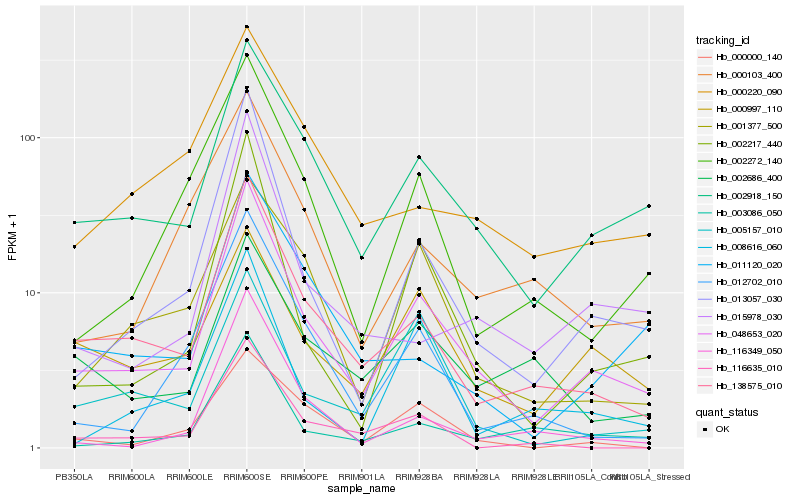
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048653_020 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_002918_150 |
0.1271060123 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 3 |
Hb_138575_010 |
0.1284032998 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_013057_030 |
0.136645427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002217_440 |
0.153049386 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_003086_050 |
0.18002406 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 7 |
Hb_002272_140 |
0.1818257314 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 8 |
Hb_000000_140 |
0.1878323722 |
- |
- |
- |
| 9 |
Hb_012702_010 |
0.189489947 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 10 |
Hb_000997_110 |
0.1905114639 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 11 |
Hb_001377_500 |
0.191531565 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 12 |
Hb_015978_030 |
0.1915684776 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 13 |
Hb_008616_060 |
0.1976841406 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 14 |
Hb_000103_400 |
0.1980178924 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000220_090 |
0.1982793272 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 16 |
Hb_011120_020 |
0.1996056874 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_116349_050 |
0.2000263906 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 18 |
Hb_005157_010 |
0.2009745256 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 19 |
Hb_002686_400 |
0.2010195676 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 20 |
Hb_116635_010 |
0.2010653253 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |