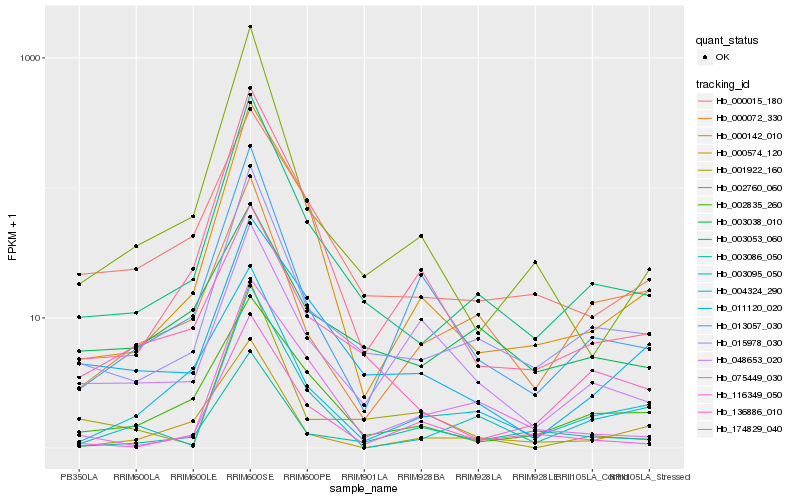
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015978_030 |
0.0 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 2 |
Hb_003053_060 |
0.0856593477 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 3 |
Hb_003086_050 |
0.1559029701 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000072_330 |
0.1700037997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004324_290 |
0.1710993283 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 6 |
Hb_013057_030 |
0.1713541255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003095_050 |
0.1776687858 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 8 |
Hb_000142_010 |
0.1793070427 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 9 |
Hb_000015_180 |
0.1798042661 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 10 |
Hb_002835_260 |
0.1798385006 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 11 |
Hb_116349_050 |
0.1826583369 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000574_120 |
0.1843509011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002760_060 |
0.1881812074 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 14 |
Hb_003038_010 |
0.1899007218 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_048653_020 |
0.1915684776 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_011120_020 |
0.193357067 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_174829_040 |
0.1964268284 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 18 |
Hb_075449_030 |
0.1967838119 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 19 |
Hb_001922_160 |
0.1989039938 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 20 |
Hb_136886_010 |
0.199681658 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |