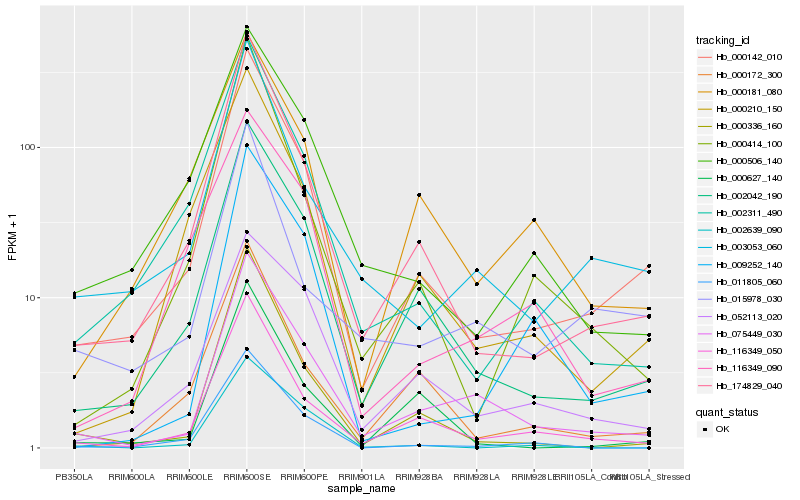
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075449_030 |
0.0 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 2 |
Hb_116349_050 |
0.1382357451 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_002042_190 |
0.1383394275 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 4 |
Hb_000142_010 |
0.1493414226 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 5 |
Hb_174829_040 |
0.1661769968 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 6 |
Hb_052113_020 |
0.1756459696 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 7 |
Hb_003053_060 |
0.1771864094 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 8 |
Hb_000336_160 |
0.1820548521 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_011805_060 |
0.1961901271 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_009252_140 |
0.1962247594 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 11 |
Hb_015978_030 |
0.1967838119 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 12 |
Hb_000172_300 |
0.2011854806 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 13 |
Hb_000181_080 |
0.2034199072 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 14 |
Hb_000210_150 |
0.2034890788 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_000506_140 |
0.2044046059 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 16 |
Hb_000627_140 |
0.2053240906 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_002311_490 |
0.2070094034 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 18 |
Hb_002639_090 |
0.2074936743 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 19 |
Hb_000414_100 |
0.207717398 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 20 |
Hb_116349_090 |
0.2078996039 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |