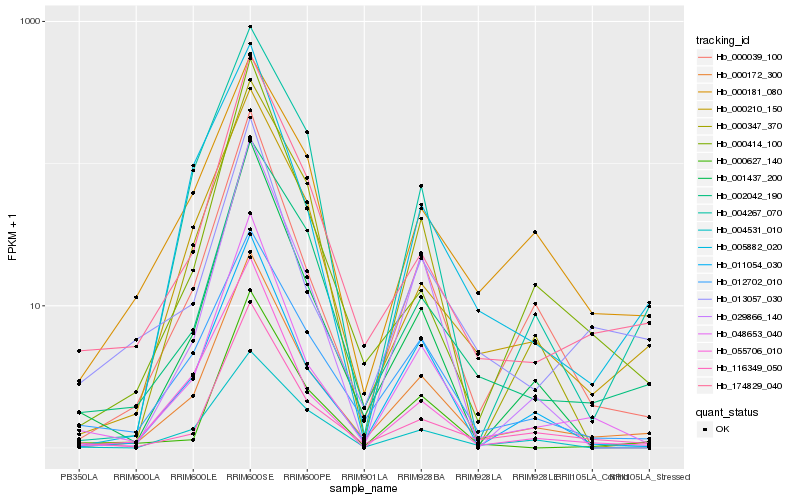
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_300 |
0.0 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 2 |
Hb_000039_100 |
0.0919805277 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 3 |
Hb_116349_050 |
0.0997260208 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_048653_040 |
0.0999758223 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 5 |
Hb_011054_030 |
0.1084273486 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 6 |
Hb_174829_040 |
0.1098303376 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 7 |
Hb_012702_010 |
0.11203182 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 8 |
Hb_000210_150 |
0.114032492 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 9 |
Hb_002042_190 |
0.1167400979 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 10 |
Hb_004267_070 |
0.1235543938 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 11 |
Hb_001437_200 |
0.1240623509 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 12 |
Hb_000347_370 |
0.1283335736 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 13 |
Hb_004531_010 |
0.1305530344 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 14 |
Hb_000181_080 |
0.130904678 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 15 |
Hb_055706_010 |
0.1311155803 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 16 |
Hb_005882_020 |
0.1388527511 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 17 |
Hb_029866_140 |
0.1396512281 |
- |
- |
PREDICTED: hevamine-A [Jatropha curcas] |
| 18 |
Hb_013057_030 |
0.1399112437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000414_100 |
0.140849657 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 20 |
Hb_000627_140 |
0.1415462438 |
- |
- |
ATP binding protein, putative [Ricinus communis] |