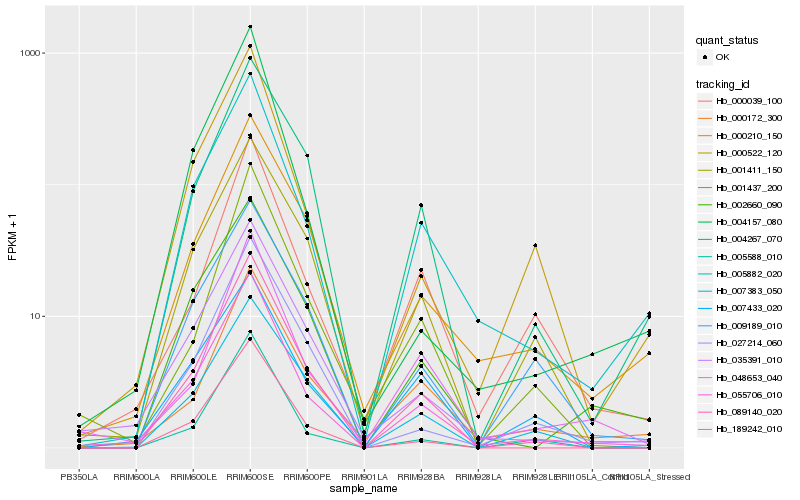
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055706_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 2 |
Hb_005882_020 |
0.0904800839 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 3 |
Hb_089140_020 |
0.0923538776 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_035391_010 |
0.1051784322 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 5 |
Hb_000522_120 |
0.1175794076 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 6 |
Hb_189242_010 |
0.1179524195 |
- |
- |
hypothetical protein B456_013G0075002, partial [Gossypium raimondii] |
| 7 |
Hb_048653_040 |
0.1216398344 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 8 |
Hb_000210_150 |
0.1235595375 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 9 |
Hb_000039_100 |
0.1286497001 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 10 |
Hb_005588_010 |
0.1286911716 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 11 |
Hb_001411_150 |
0.1306197052 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 12 |
Hb_000172_300 |
0.1311155803 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 13 |
Hb_002660_090 |
0.1335202093 |
- |
- |
hypothetical protein JCGZ_16500 [Jatropha curcas] |
| 14 |
Hb_001437_200 |
0.1354155553 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 15 |
Hb_004267_070 |
0.1355097333 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 16 |
Hb_007433_020 |
0.1390961737 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 17 |
Hb_007383_050 |
0.1408381517 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 18 |
Hb_027214_060 |
0.1415418242 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 19 |
Hb_009189_010 |
0.1424422244 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 20 |
Hb_004157_080 |
0.1436134306 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |