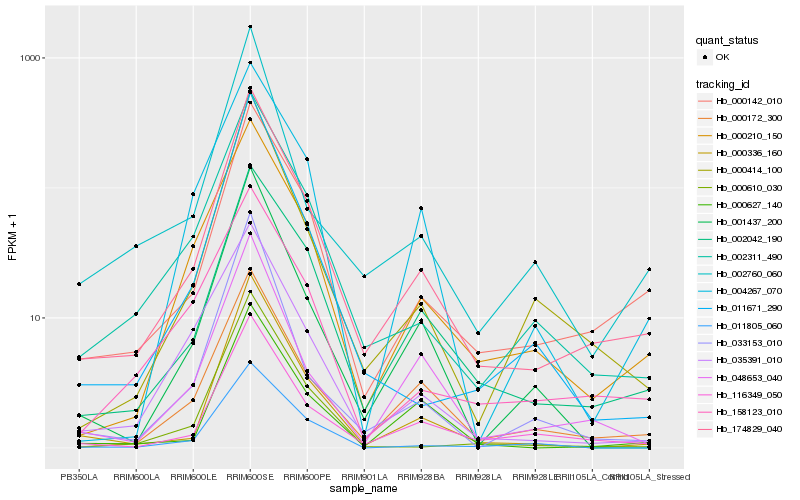
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174829_040 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 2 |
Hb_000142_010 |
0.08297079 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 3 |
Hb_002042_190 |
0.0925368157 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 4 |
Hb_116349_050 |
0.1030381804 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_002311_490 |
0.1041833489 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 6 |
Hb_000172_300 |
0.1098303376 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 7 |
Hb_000414_100 |
0.1113589484 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 8 |
Hb_000336_160 |
0.1119142273 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000210_150 |
0.1143041541 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 10 |
Hb_001437_200 |
0.1289167163 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 11 |
Hb_011671_290 |
0.1372305357 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 12 |
Hb_000627_140 |
0.1390066164 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_048653_040 |
0.1391460577 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 14 |
Hb_035391_010 |
0.1399138913 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 15 |
Hb_000610_030 |
0.1410829831 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |
| 16 |
Hb_158123_010 |
0.1415860695 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 17 |
Hb_011805_060 |
0.1464120296 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 18 |
Hb_002760_060 |
0.146747653 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 19 |
Hb_004267_070 |
0.1482710227 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 20 |
Hb_033153_010 |
0.1494937837 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |