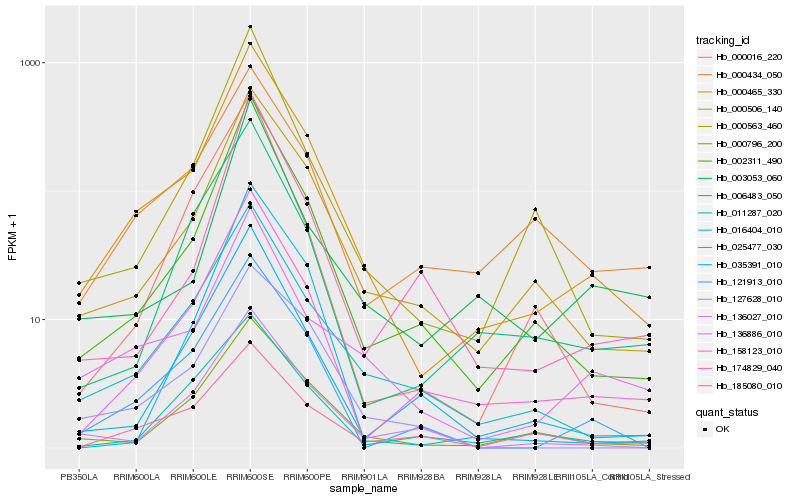
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_330 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 2 |
Hb_002311_490 |
0.1073964605 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 3 |
Hb_000506_140 |
0.1115365644 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 4 |
Hb_158123_010 |
0.1173445293 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_016404_010 |
0.119716962 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_000796_200 |
0.1310879176 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 7 |
Hb_121913_010 |
0.1374636341 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 8 |
Hb_000563_460 |
0.1455665808 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 9 |
Hb_136886_010 |
0.146122745 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 10 |
Hb_006483_050 |
0.1556607858 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 11 |
Hb_136027_010 |
0.1616924343 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_127628_010 |
0.1619864926 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 13 |
Hb_000434_050 |
0.1635718436 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 14 |
Hb_035391_010 |
0.1636068247 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 15 |
Hb_185080_010 |
0.1671879765 |
- |
- |
- |
| 16 |
Hb_011287_020 |
0.1700980237 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_025477_030 |
0.1721201807 |
transcription factor |
TF Family: ERF |
dehydration-responsive element-binding protein 2 [Manihot esculenta] |
| 18 |
Hb_000016_220 |
0.1742496035 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 19 |
Hb_003053_060 |
0.1767453745 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 20 |
Hb_174829_040 |
0.1775849615 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |