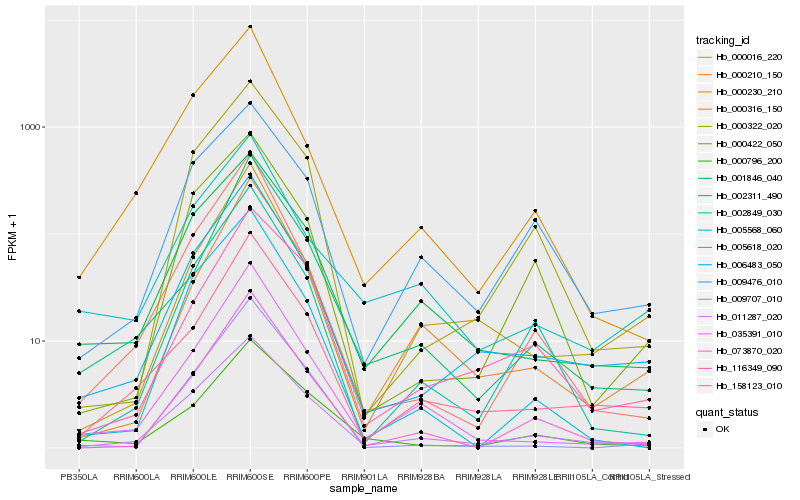
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006483_050 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 2 |
Hb_158123_010 |
0.0935660385 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_011287_020 |
0.0947466286 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000316_150 |
0.1120820051 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 5 |
Hb_000016_220 |
0.1172651024 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 6 |
Hb_001846_040 |
0.1182811377 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_000322_020 |
0.1274829535 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 8 |
Hb_002849_030 |
0.1301355092 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 9 |
Hb_005568_060 |
0.1329779683 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |
| 10 |
Hb_000210_150 |
0.1334695866 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 11 |
Hb_035391_010 |
0.1337636021 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 12 |
Hb_000422_050 |
0.1341656487 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 13 |
Hb_073870_020 |
0.1345847504 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 14 |
Hb_000230_210 |
0.1358276217 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 15 |
Hb_005618_020 |
0.1390901776 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 16 |
Hb_000796_200 |
0.1415918898 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 17 |
Hb_009476_010 |
0.1420030433 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 18 |
Hb_116349_090 |
0.1422346972 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 19 |
Hb_009707_010 |
0.1432996544 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 20 |
Hb_002311_490 |
0.1435170421 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |