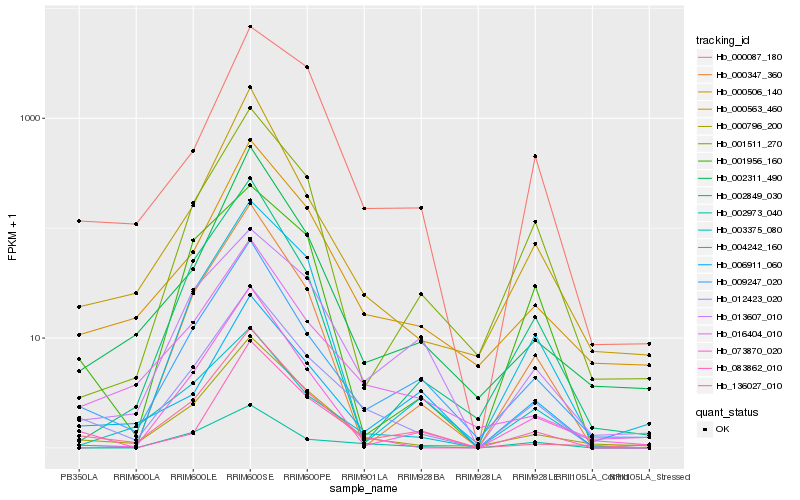
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012423_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |
| 2 |
Hb_000796_200 |
0.1054652192 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 3 |
Hb_009247_020 |
0.1273075794 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 4 |
Hb_136027_010 |
0.1369791406 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_000506_140 |
0.1385967842 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 6 |
Hb_016404_010 |
0.1402253222 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 7 |
Hb_000563_460 |
0.1540411704 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 8 |
Hb_002973_040 |
0.1566787697 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 9 |
Hb_083862_010 |
0.1570549696 |
- |
- |
hypothetical protein JCGZ_17956 [Jatropha curcas] |
| 10 |
Hb_003375_080 |
0.1578879066 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001511_270 |
0.1631646112 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 12 |
Hb_002311_490 |
0.1685339289 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 13 |
Hb_000347_360 |
0.1708526472 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002849_030 |
0.1709643533 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 15 |
Hb_000087_180 |
0.1714653229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_073870_020 |
0.1725338451 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 17 |
Hb_006911_060 |
0.1744146208 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |
| 18 |
Hb_004242_160 |
0.1756254476 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001956_160 |
0.1774482495 |
- |
- |
hypothetical protein L484_009438 [Morus notabilis] |
| 20 |
Hb_013607_010 |
0.1782637359 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |