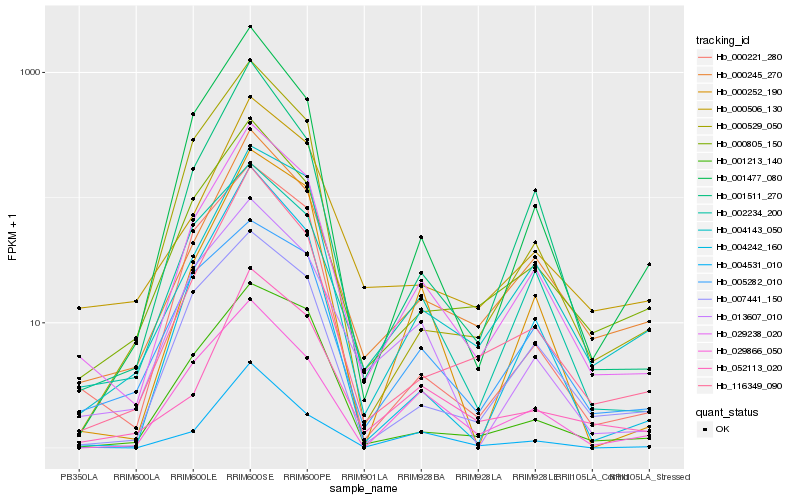
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029238_020 |
0.0 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 2 |
Hb_000245_270 |
0.0981743135 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 3 |
Hb_116349_090 |
0.1063056301 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 4 |
Hb_000805_150 |
0.1148213487 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_001511_270 |
0.1152323717 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 6 |
Hb_002234_200 |
0.1197012319 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 7 |
Hb_000506_130 |
0.12041135 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 8 |
Hb_004143_050 |
0.12148685 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 9 |
Hb_000252_190 |
0.1247298033 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 10 |
Hb_001477_080 |
0.1248934947 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 11 |
Hb_007441_150 |
0.1261223712 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 12 |
Hb_004242_160 |
0.1265515946 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000221_280 |
0.1279727732 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 14 |
Hb_013607_010 |
0.128413533 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 15 |
Hb_004531_010 |
0.134928638 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 16 |
Hb_001213_140 |
0.135808002 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 17 |
Hb_005282_010 |
0.1373878569 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 18 |
Hb_052113_020 |
0.1374675951 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 19 |
Hb_000529_050 |
0.1379436801 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 20 |
Hb_029866_050 |
0.139187428 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |