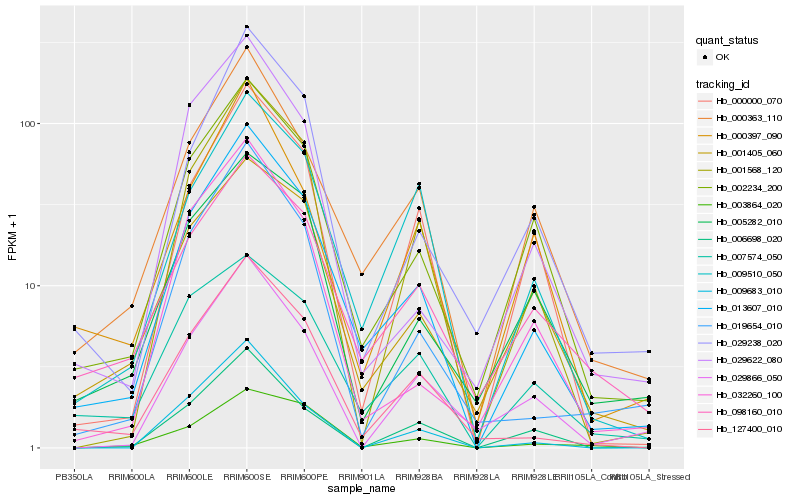
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013607_010 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 2 |
Hb_002234_200 |
0.1064341025 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 3 |
Hb_000363_110 |
0.1075658533 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 4 |
Hb_127400_010 |
0.1118102758 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_001568_120 |
0.1209963892 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 6 |
Hb_029238_020 |
0.128413533 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 7 |
Hb_009510_050 |
0.1311370437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_019654_010 |
0.133343743 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 9 |
Hb_098160_010 |
0.1345915062 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001405_060 |
0.1358126224 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 11 |
Hb_000000_070 |
0.1365387226 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 12 |
Hb_032260_100 |
0.1425539291 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 13 |
Hb_007574_050 |
0.1450878684 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 14 |
Hb_006698_020 |
0.1452596163 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 15 |
Hb_029622_080 |
0.1456530916 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 16 |
Hb_000397_090 |
0.1479833429 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 17 |
Hb_009683_010 |
0.1481400559 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_005282_010 |
0.1489081276 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 19 |
Hb_029866_050 |
0.1500081845 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 20 |
Hb_003864_020 |
0.1513853099 |
- |
- |
kinase, putative [Ricinus communis] |