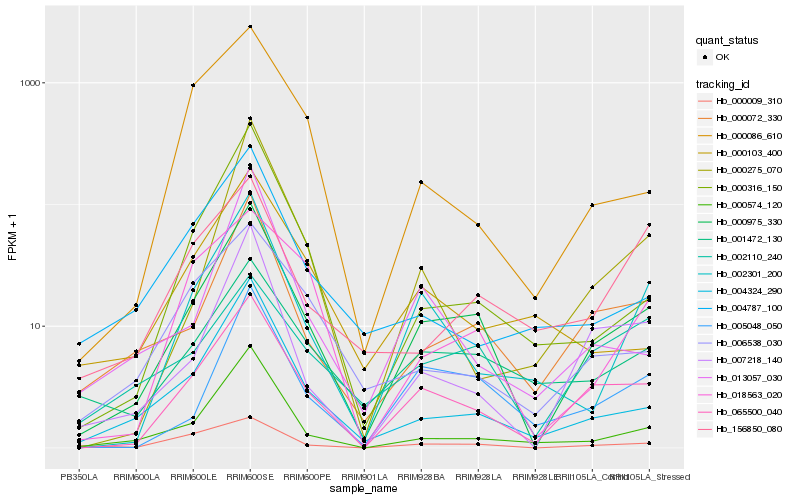
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_330 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |
| 2 |
Hb_065500_040 |
0.1205502968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000072_330 |
0.1659857125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004324_290 |
0.1742195137 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 5 |
Hb_005048_050 |
0.1812911377 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 6 |
Hb_000574_120 |
0.1814503057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000009_310 |
0.1831543573 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_007218_140 |
0.1929597262 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 9 |
Hb_001472_130 |
0.1942930111 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 10 |
Hb_002301_200 |
0.1947743598 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 11 |
Hb_000316_150 |
0.2028628014 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_018563_020 |
0.2206700481 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 6 [Hevea brasiliensis] |
| 13 |
Hb_006538_030 |
0.2213692519 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 14 |
Hb_000086_610 |
0.225390582 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 15 |
Hb_002110_240 |
0.2323864847 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 16 |
Hb_013057_030 |
0.2328101408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000103_400 |
0.2335306247 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000275_070 |
0.2340705372 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 19 |
Hb_004787_100 |
0.2364268435 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 20 |
Hb_156850_080 |
0.2404213948 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |