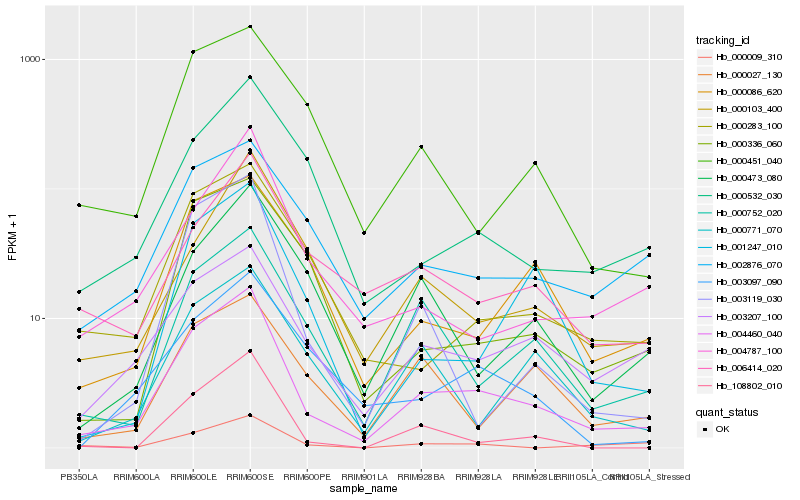
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004460_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000752_020 |
0.1818051995 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003207_100 |
0.1838651151 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 4 |
Hb_002876_070 |
0.1942780421 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 5 |
Hb_000283_100 |
0.1966033307 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_108802_010 |
0.1991110776 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000086_620 |
0.2010996747 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003119_030 |
0.2011802043 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Jatropha curcas] |
| 9 |
Hb_004787_100 |
0.2041656176 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 10 |
Hb_000532_030 |
0.2050314455 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 11 |
Hb_000103_400 |
0.2050372781 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000451_040 |
0.205306908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006414_020 |
0.2059105347 |
transcription factor |
TF Family: MYB |
- |
| 14 |
Hb_000009_310 |
0.2064973214 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000771_070 |
0.206650872 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001247_010 |
0.2080116184 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 17 |
Hb_000473_080 |
0.211148611 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 18 |
Hb_000336_060 |
0.2129243789 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 19 |
Hb_003097_090 |
0.2135431253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000027_130 |
0.2164279411 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |