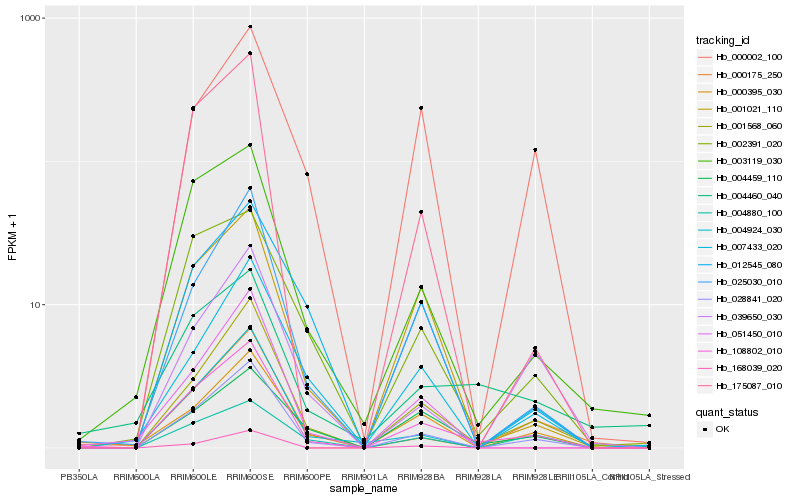
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108802_010 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 2 |
Hb_028841_020 |
0.1073883257 |
- |
- |
hypothetical protein CISIN_1g041345mg [Citrus sinensis] |
| 3 |
Hb_001568_060 |
0.1339815277 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 4 |
Hb_003119_030 |
0.1503410116 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Jatropha curcas] |
| 5 |
Hb_025030_010 |
0.150694498 |
- |
- |
- |
| 6 |
Hb_175087_010 |
0.1668202139 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 7 |
Hb_001021_110 |
0.1669737743 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 8 |
Hb_002391_020 |
0.1757542406 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 9 |
Hb_000395_030 |
0.176481303 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 10 |
Hb_000175_250 |
0.1802389665 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 11 |
Hb_051450_010 |
0.1845733964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004924_030 |
0.1955864269 |
- |
- |
- |
| 13 |
Hb_007433_020 |
0.1967968098 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 14 |
Hb_039650_030 |
0.1982838658 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 15 |
Hb_004460_040 |
0.1991110776 |
- |
- |
- |
| 16 |
Hb_168039_020 |
0.2011054089 |
- |
- |
- |
| 17 |
Hb_004880_100 |
0.2013758965 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |
| 18 |
Hb_012545_080 |
0.2041566306 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 19 |
Hb_004459_110 |
0.2043243603 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 20 |
Hb_000002_100 |
0.2056757253 |
- |
- |
phosphate transporter [Manihot esculenta] |