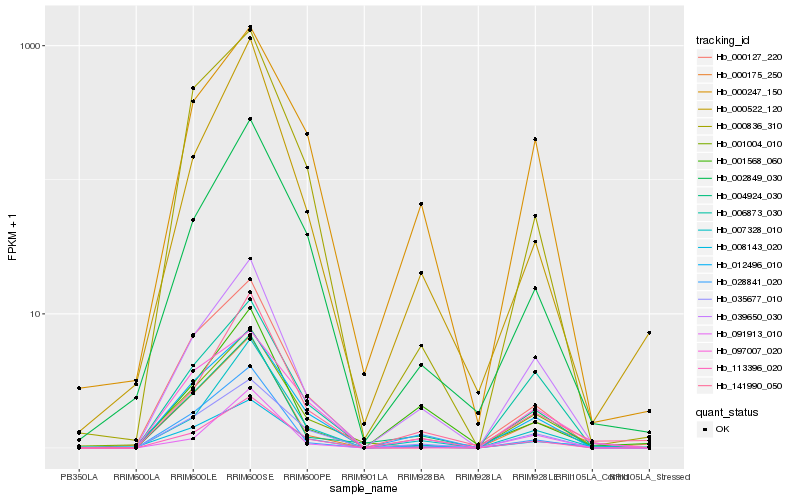
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_250 |
0.0 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 2 |
Hb_039650_030 |
0.0799867685 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 3 |
Hb_004924_030 |
0.1166010097 |
- |
- |
- |
| 4 |
Hb_028841_020 |
0.1284696403 |
- |
- |
hypothetical protein CISIN_1g041345mg [Citrus sinensis] |
| 5 |
Hb_007328_010 |
0.1395899354 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_012496_010 |
0.1415799103 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_141990_050 |
0.1423004434 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 8 |
Hb_035677_010 |
0.1484941798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001004_010 |
0.1507742858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001568_060 |
0.1516877682 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 11 |
Hb_008143_020 |
0.1544550692 |
- |
- |
PREDICTED: rhomboid-like protease 2 [Jatropha curcas] |
| 12 |
Hb_000247_150 |
0.1553388455 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 13 |
Hb_097007_020 |
0.1592358081 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 14 |
Hb_000836_310 |
0.1599747215 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 30 [Jatropha curcas] |
| 15 |
Hb_000127_220 |
0.1636470776 |
- |
- |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 16 |
Hb_113396_020 |
0.1639201085 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 17 |
Hb_006873_030 |
0.1659925209 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 18 |
Hb_000522_120 |
0.1669567413 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 19 |
Hb_002849_030 |
0.1731752534 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 20 |
Hb_091913_010 |
0.1736419429 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |