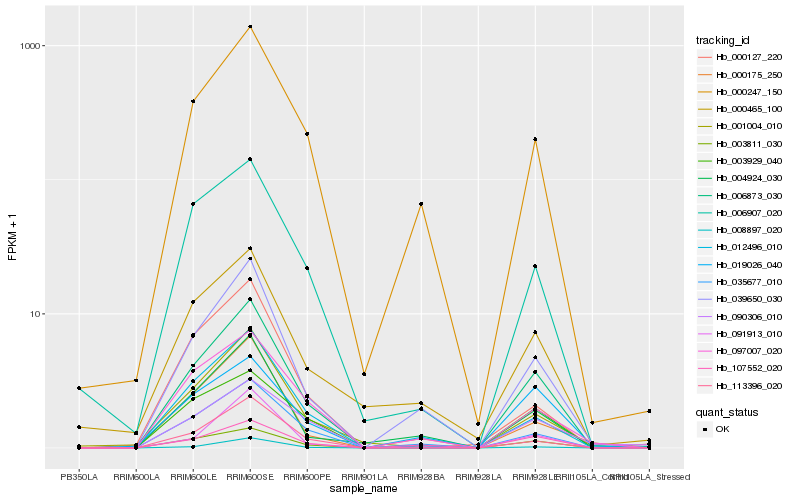
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006873_030 |
0.0 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_003811_030 |
0.1034136717 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 3 |
Hb_012496_010 |
0.1155970236 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_039650_030 |
0.1213414095 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 5 |
Hb_113396_020 |
0.1295805374 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 6 |
Hb_035677_010 |
0.130605074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000247_150 |
0.1378772542 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_001004_010 |
0.137979551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003929_040 |
0.1391294321 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_006907_020 |
0.1442881485 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase 3 [Manihot esculenta] |
| 11 |
Hb_107552_020 |
0.1505390755 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 12 |
Hb_004924_030 |
0.1506912701 |
- |
- |
- |
| 13 |
Hb_097007_020 |
0.1544538815 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 14 |
Hb_091913_010 |
0.1572975176 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 15 |
Hb_008897_020 |
0.1635054425 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Musa acuminata subsp. malaccensis] |
| 16 |
Hb_000175_250 |
0.1659925209 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 17 |
Hb_019026_040 |
0.1669506318 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 18 |
Hb_090306_010 |
0.1699814837 |
- |
- |
hypothetical protein POPTR_0007s02300g [Populus trichocarpa] |
| 19 |
Hb_000127_220 |
0.1705751049 |
- |
- |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 20 |
Hb_000465_100 |
0.1716382196 |
- |
- |
catalytic, putative [Ricinus communis] |