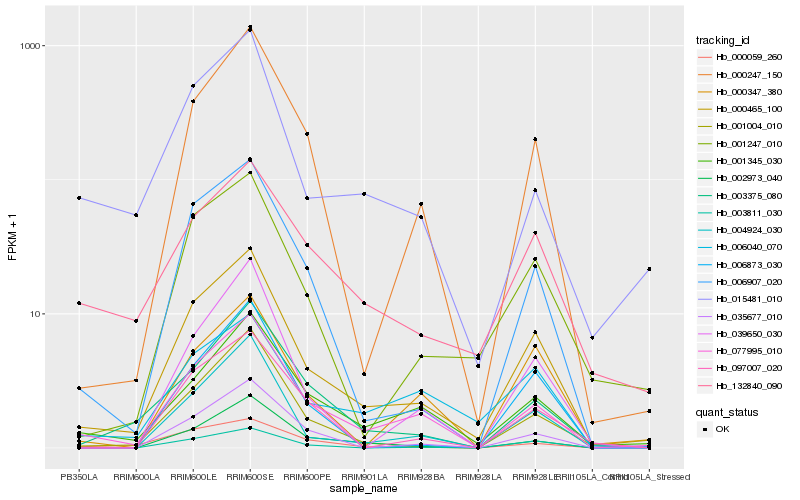
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_100 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_077995_010 |
0.1159751849 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 3 |
Hb_001247_010 |
0.141450702 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 4 |
Hb_006907_020 |
0.1420289917 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase 3 [Manihot esculenta] |
| 5 |
Hb_004924_030 |
0.1467331439 |
- |
- |
- |
| 6 |
Hb_001345_030 |
0.1471638783 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 7 |
Hb_000247_150 |
0.1472350715 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_001004_010 |
0.1477561956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003811_030 |
0.1544694573 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 10 |
Hb_003375_080 |
0.1635099542 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 11 |
Hb_039650_030 |
0.1670158631 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 12 |
Hb_002973_040 |
0.1693401203 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 13 |
Hb_000347_380 |
0.1710485006 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_035677_010 |
0.1711194496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006873_030 |
0.1716382196 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 16 |
Hb_132840_090 |
0.1740010127 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_097007_020 |
0.1750499763 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 18 |
Hb_015481_010 |
0.1789969981 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 19 |
Hb_000059_260 |
0.1792767779 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: peroxidase 60 [Jatropha curcas] |
| 20 |
Hb_006040_070 |
0.1794136762 |
- |
- |
orf107b gene product (mitochondrion) [Beta vulgaris subsp. vulgaris] |