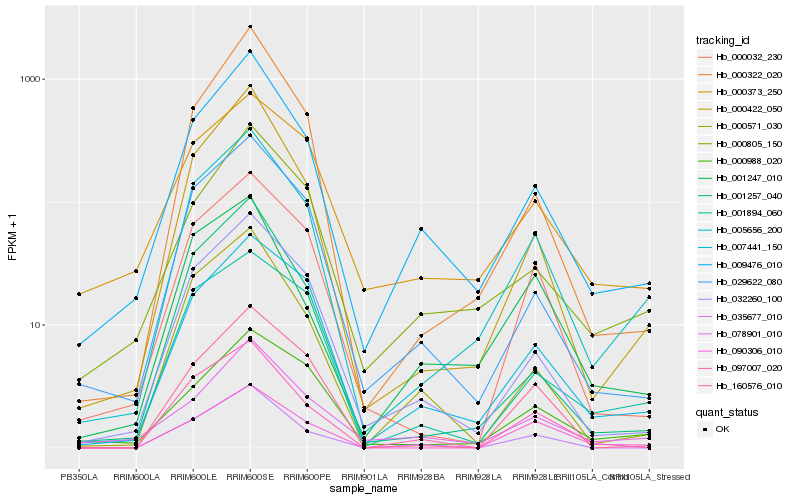
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005656_200 |
0.0 |
- |
- |
Cation transport protein chaC, putative [Ricinus communis] |
| 2 |
Hb_000422_050 |
0.1196309573 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 3 |
Hb_009476_010 |
0.1196461965 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 4 |
Hb_007441_150 |
0.1196554664 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 5 |
Hb_000032_230 |
0.1271417697 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20463 [Jatropha curcas] |
| 6 |
Hb_001894_060 |
0.1354543999 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 7 |
Hb_032260_100 |
0.1375142779 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 8 |
Hb_000805_150 |
0.1440666713 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000988_020 |
0.1443391368 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 10 |
Hb_029622_080 |
0.1451237035 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 11 |
Hb_090306_010 |
0.1451295392 |
- |
- |
hypothetical protein POPTR_0007s02300g [Populus trichocarpa] |
| 12 |
Hb_097007_020 |
0.1503388957 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 13 |
Hb_160576_010 |
0.1511303567 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000322_020 |
0.1537492779 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 15 |
Hb_001247_010 |
0.1542172215 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 16 |
Hb_000571_030 |
0.1550475682 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 17 |
Hb_078901_010 |
0.1562738895 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 18 |
Hb_001257_040 |
0.1579393433 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 19 |
Hb_000373_250 |
0.162193347 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 20 |
Hb_035677_010 |
0.1636040128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |