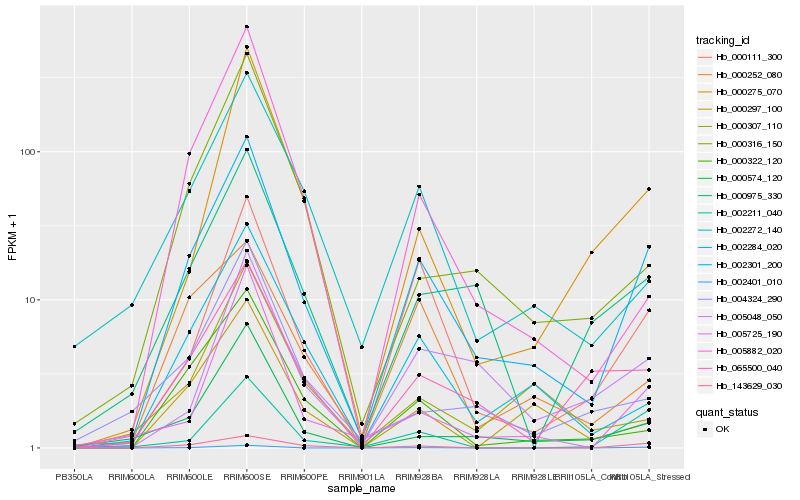
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_200 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 2 |
Hb_002284_020 |
0.1831144915 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 3 |
Hb_000111_300 |
0.1840273846 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 4 |
Hb_143629_030 |
0.1898914455 |
- |
- |
hypothetical protein JCGZ_23143 [Jatropha curcas] |
| 5 |
Hb_000574_120 |
0.1929192708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000975_330 |
0.1947743598 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |
| 7 |
Hb_065500_040 |
0.1953938198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000322_120 |
0.1997012443 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 9 |
Hb_000275_070 |
0.2052628779 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 10 |
Hb_005725_190 |
0.2072168674 |
- |
- |
hypothetical protein JCGZ_15992 [Jatropha curcas] |
| 11 |
Hb_002272_140 |
0.2078056532 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 12 |
Hb_000297_100 |
0.2079387765 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 13 |
Hb_000316_150 |
0.2126687171 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_005882_020 |
0.2128458154 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 15 |
Hb_005048_050 |
0.2136272307 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 16 |
Hb_002401_010 |
0.2185120211 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 17 |
Hb_002211_040 |
0.2185464706 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 18 |
Hb_000252_080 |
0.2201083371 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 19 |
Hb_004324_290 |
0.2284574171 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 20 |
Hb_000307_110 |
0.2294130972 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |