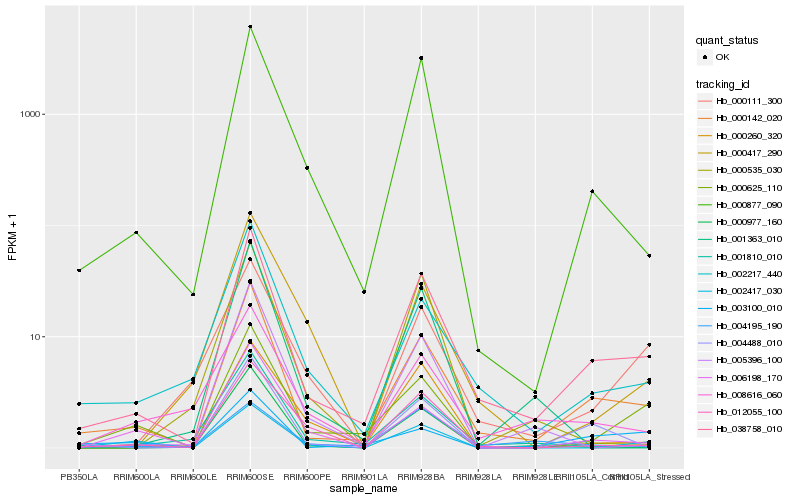
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038758_010 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000142_020 |
0.0656437185 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000877_090 |
0.1605152987 |
- |
- |
allergenic-related protein Pt2L4 [Manihot esculenta] |
| 4 |
Hb_002217_440 |
0.1768917252 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000625_110 |
0.1863103896 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001810_010 |
0.1889037911 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 7 |
Hb_012055_100 |
0.1976540677 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 8 |
Hb_008616_060 |
0.2005349064 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 9 |
Hb_000260_320 |
0.2027587465 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 10 |
Hb_000111_300 |
0.2035053844 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 11 |
Hb_004488_010 |
0.2045059674 |
- |
- |
hypothetical protein CISIN_1g019168mg [Citrus sinensis] |
| 12 |
Hb_000977_160 |
0.2112464733 |
- |
- |
Serine-threonine protein kinase, plant-type, putative [Theobroma cacao] |
| 13 |
Hb_005396_100 |
0.2157636854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004195_190 |
0.222234595 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_001363_010 |
0.2226849355 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 16 |
Hb_002417_030 |
0.224036171 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 17 |
Hb_006198_170 |
0.2284067398 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070-like [Jatropha curcas] |
| 18 |
Hb_003100_010 |
0.2303411736 |
- |
- |
PREDICTED: uncharacterized protein LOC101217345 [Cucumis sativus] |
| 19 |
Hb_000535_030 |
0.2303827554 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 20 |
Hb_000417_290 |
0.231608463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |