| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
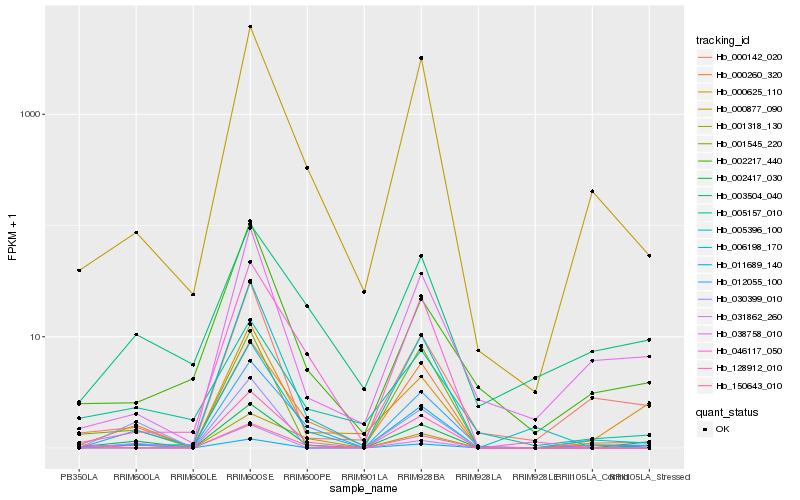
Hb_002417_030 |
0.0 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 2 |
Hb_000625_110 |
0.1570141077 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 3 |
Hb_012055_100 |
0.2186979601 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 4 |
Hb_006198_170 |
0.222844605 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070-like [Jatropha curcas] |
| 5 |
Hb_038758_010 |
0.224036171 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000877_090 |
0.2241475094 |
- |
- |
allergenic-related protein Pt2L4 [Manihot esculenta] |
| 7 |
Hb_000142_020 |
0.2275874067 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000260_320 |
0.2314447991 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 9 |
Hb_001318_130 |
0.2360869781 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 10 |
Hb_030399_010 |
0.2362361877 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 11 |
Hb_031862_260 |
0.2381527414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011689_140 |
0.2425126907 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 4 [Jatropha curcas] |
| 13 |
Hb_001545_220 |
0.2428281624 |
- |
- |
hypothetical protein POPTR_0002s06170g [Populus trichocarpa] |
| 14 |
Hb_005157_010 |
0.2477502965 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 15 |
Hb_003504_040 |
0.2481678869 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 16 |
Hb_128912_010 |
0.2492935569 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 17 |
Hb_002217_440 |
0.2559533308 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_046117_050 |
0.2567122695 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 19 |
Hb_005396_100 |
0.2577552085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_150643_010 |
0.2603202727 |
- |
- |
lipid binding protein, putative [Ricinus communis] |