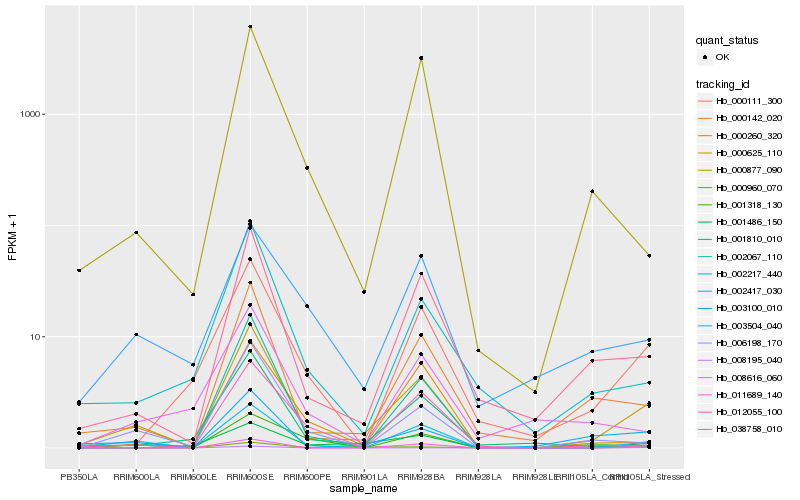
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000625_110 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002417_030 |
0.1570141077 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 3 |
Hb_038758_010 |
0.1863103896 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000142_020 |
0.1884599011 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_003100_010 |
0.2054486773 |
- |
- |
PREDICTED: uncharacterized protein LOC101217345 [Cucumis sativus] |
| 6 |
Hb_006198_170 |
0.213465825 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070-like [Jatropha curcas] |
| 7 |
Hb_011689_140 |
0.2255920082 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 4 [Jatropha curcas] |
| 8 |
Hb_002217_440 |
0.2268210664 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000960_070 |
0.2334601864 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 10 |
Hb_000111_300 |
0.2336134522 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 11 |
Hb_001318_130 |
0.2393396842 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 12 |
Hb_000877_090 |
0.2410539608 |
- |
- |
allergenic-related protein Pt2L4 [Manihot esculenta] |
| 13 |
Hb_003504_040 |
0.2483458467 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 14 |
Hb_012055_100 |
0.2493002657 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 15 |
Hb_001810_010 |
0.250402206 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 16 |
Hb_000260_320 |
0.2547633944 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 17 |
Hb_001486_150 |
0.255192224 |
- |
- |
- |
| 18 |
Hb_008195_040 |
0.2590510218 |
- |
- |
hypothetical protein VITISV_033788 [Vitis vinifera] |
| 19 |
Hb_008616_060 |
0.262694605 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 20 |
Hb_002067_110 |
0.262876155 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |