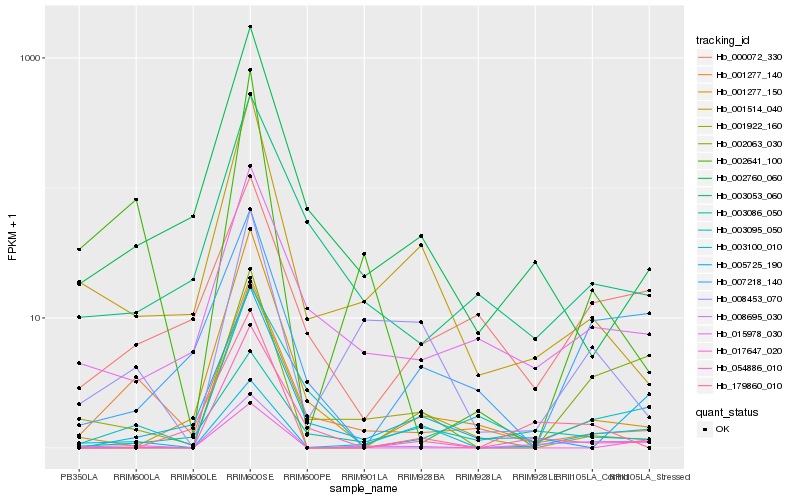
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008695_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_12043 [Jatropha curcas] |
| 2 |
Hb_002063_030 |
0.2323116608 |
- |
- |
- |
| 3 |
Hb_003100_010 |
0.2507234361 |
- |
- |
PREDICTED: uncharacterized protein LOC101217345 [Cucumis sativus] |
| 4 |
Hb_015978_030 |
0.2527011133 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 5 |
Hb_001514_040 |
0.2682690721 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 6 |
Hb_054886_010 |
0.2710828877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_179860_010 |
0.2716402738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002760_060 |
0.2753253134 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 9 |
Hb_005725_190 |
0.2768852925 |
- |
- |
hypothetical protein JCGZ_15992 [Jatropha curcas] |
| 10 |
Hb_002641_100 |
0.2776066955 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 11 |
Hb_008453_070 |
0.2783726637 |
- |
- |
putative aquaporin [Vitis vinifera] |
| 12 |
Hb_003086_050 |
0.2785510599 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 13 |
Hb_001922_160 |
0.2820774462 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 14 |
Hb_007218_140 |
0.282850706 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 15 |
Hb_003095_050 |
0.2844903425 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 16 |
Hb_001277_140 |
0.2848445423 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |
| 17 |
Hb_017647_020 |
0.2863995991 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 18 |
Hb_001277_150 |
0.2866607925 |
- |
- |
hypothetical protein VITISV_043492 [Vitis vinifera] |
| 19 |
Hb_003053_060 |
0.2872022491 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 20 |
Hb_000072_330 |
0.2877243713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |