| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
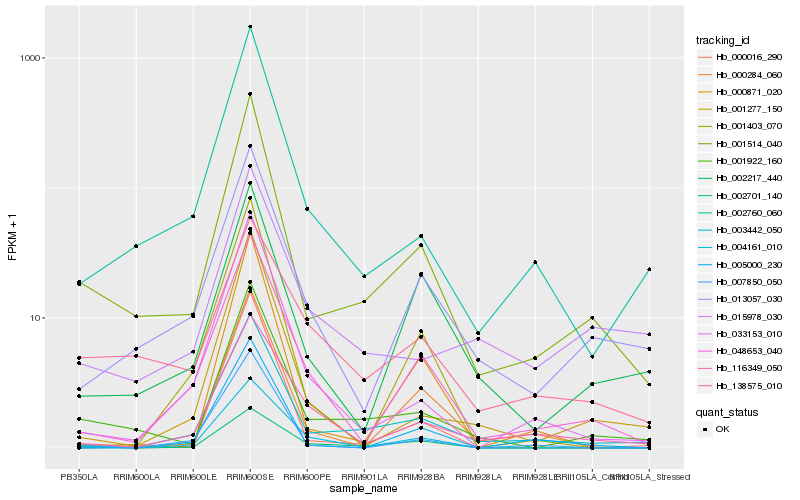
Hb_001514_040 |
0.0 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 2 |
Hb_001922_160 |
0.1103815278 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_002760_060 |
0.1494073993 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 4 |
Hb_003442_050 |
0.1591927327 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 5 |
Hb_013057_030 |
0.1618320221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_048653_040 |
0.1758521338 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 7 |
Hb_033153_010 |
0.1824057308 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 8 |
Hb_001277_150 |
0.188134912 |
- |
- |
hypothetical protein VITISV_043492 [Vitis vinifera] |
| 9 |
Hb_002217_440 |
0.1886352914 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_007850_050 |
0.1913404532 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_002701_140 |
0.2018276917 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 12 |
Hb_005000_230 |
0.202846976 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 13 |
Hb_116349_050 |
0.2050709009 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_001403_070 |
0.2061401065 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 15 |
Hb_015978_030 |
0.2070285453 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 16 |
Hb_000871_020 |
0.2075783452 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 17 |
Hb_000284_060 |
0.2080150816 |
desease resistance |
Gene Name: NB-ARC |
disease resistance protein (TIR-NBS-LRR class), putative [Medicago truncatula] |
| 18 |
Hb_000016_290 |
0.208055818 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 19 |
Hb_004161_010 |
0.2082999234 |
- |
- |
hypothetical protein JCGZ_21278 [Jatropha curcas] |
| 20 |
Hb_138575_010 |
0.2095342036 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |