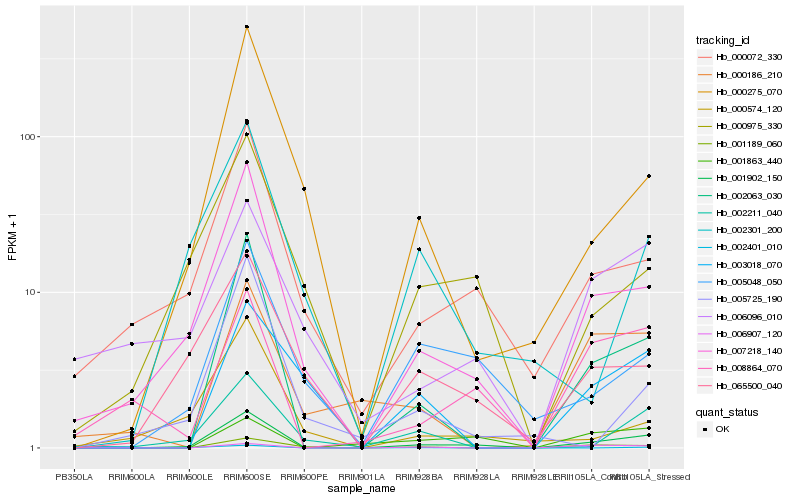
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001902_150 |
0.0 |
- |
- |
PREDICTED: cytokinin hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_007218_140 |
0.1651470031 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 3 |
Hb_002063_030 |
0.1976482485 |
- |
- |
- |
| 4 |
Hb_000072_330 |
0.2314977641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005048_050 |
0.2377257285 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 6 |
Hb_000275_070 |
0.2457053506 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 7 |
Hb_001189_060 |
0.2470052452 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 8 |
Hb_065500_040 |
0.2510690099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000975_330 |
0.2566522084 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |
| 10 |
Hb_008864_070 |
0.2656186772 |
- |
- |
PREDICTED: uncharacterized protein LOC105803851 [Gossypium raimondii] |
| 11 |
Hb_002211_040 |
0.2673822543 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 12 |
Hb_003018_070 |
0.2836808023 |
- |
- |
SAUR family protein, partial [Theobroma cacao] |
| 13 |
Hb_002401_010 |
0.2849264763 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 14 |
Hb_006096_010 |
0.2855216079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002301_200 |
0.2880058074 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 16 |
Hb_000574_120 |
0.2885367478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005725_190 |
0.295051998 |
- |
- |
hypothetical protein JCGZ_15992 [Jatropha curcas] |
| 18 |
Hb_006907_120 |
0.2982104295 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 19 |
Hb_001863_440 |
0.299190976 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 20 |
Hb_000186_210 |
0.2996468921 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |