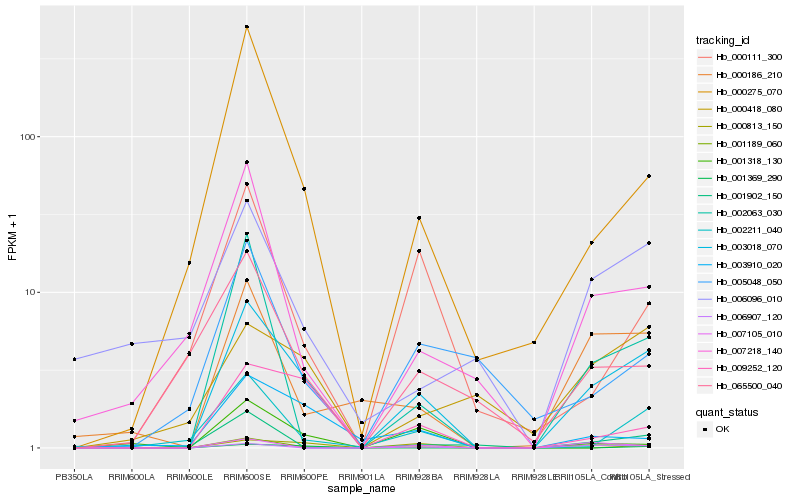
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003018_070 |
0.0 |
- |
- |
SAUR family protein, partial [Theobroma cacao] |
| 2 |
Hb_001189_060 |
0.1554314983 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 3 |
Hb_000186_210 |
0.2704392876 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 4 |
Hb_000275_070 |
0.278407176 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 5 |
Hb_009252_120 |
0.2827899059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001902_150 |
0.2836808023 |
- |
- |
PREDICTED: cytokinin hydroxylase-like [Jatropha curcas] |
| 7 |
Hb_002211_040 |
0.283932068 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 8 |
Hb_000418_080 |
0.2988337542 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 9 |
Hb_006907_120 |
0.2999403012 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 10 |
Hb_002063_030 |
0.3029659017 |
- |
- |
- |
| 11 |
Hb_003910_020 |
0.3058197736 |
- |
- |
PREDICTED: reticulon-like protein B13 [Jatropha curcas] |
| 12 |
Hb_007218_140 |
0.3100502972 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 13 |
Hb_000813_150 |
0.313339528 |
- |
- |
- |
| 14 |
Hb_065500_040 |
0.3166610463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001318_130 |
0.3198032488 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 16 |
Hb_000111_300 |
0.3248538712 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 17 |
Hb_005048_050 |
0.3253831827 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 18 |
Hb_006096_010 |
0.3264082839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007105_010 |
0.3278798813 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 20 |
Hb_001369_290 |
0.3291683071 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |