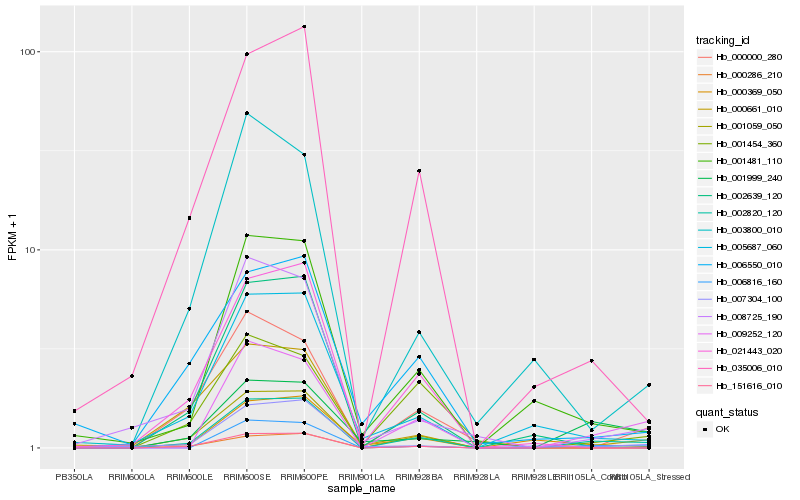
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002820_120 |
0.0 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 2 |
Hb_009252_120 |
0.1589152572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006816_160 |
0.1612611315 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 4 |
Hb_035006_010 |
0.1907327337 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 5 |
Hb_001481_110 |
0.1967077612 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 6 |
Hb_005687_060 |
0.1973354541 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001059_050 |
0.2066873542 |
- |
- |
PREDICTED: putative F-box/LRR-repeat protein At3g18150 [Jatropha curcas] |
| 8 |
Hb_000000_280 |
0.2071276635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008725_190 |
0.2086282883 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 10 |
Hb_001999_240 |
0.2086393622 |
- |
- |
PREDICTED: josephin-like protein [Populus euphratica] |
| 11 |
Hb_151616_010 |
0.2097859918 |
- |
- |
- |
| 12 |
Hb_000286_210 |
0.2124425487 |
- |
- |
hypothetical protein EUGRSUZ_C00018 [Eucalyptus grandis] |
| 13 |
Hb_001454_360 |
0.2145172369 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 14 |
Hb_021443_020 |
0.216567586 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 15 |
Hb_007304_100 |
0.2176952263 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 16 |
Hb_002639_120 |
0.217907135 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000661_010 |
0.2194130569 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 18 |
Hb_000369_050 |
0.2198801814 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 19 |
Hb_003800_010 |
0.2245456597 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 20 |
Hb_006550_010 |
0.2250648325 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |