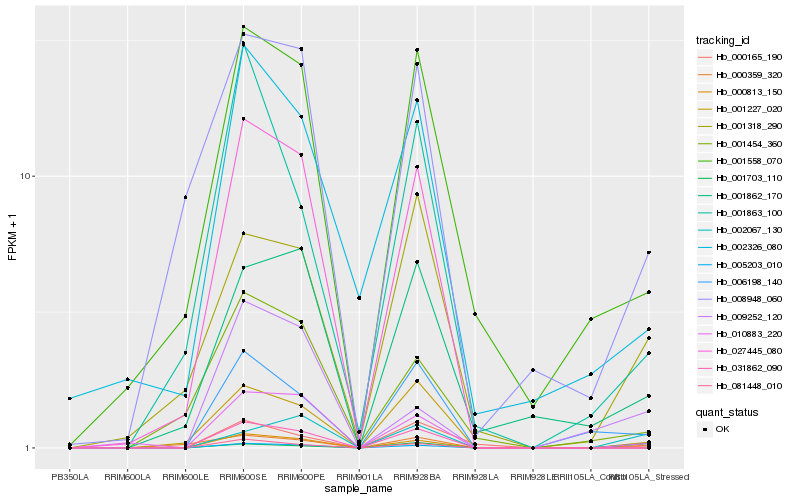
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000813_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_002067_130 |
0.2162209139 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |
| 3 |
Hb_010883_220 |
0.2237584035 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Prunus mume] |
| 4 |
Hb_009252_120 |
0.2288387545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001703_110 |
0.23301545 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 6 |
Hb_006198_140 |
0.2426434216 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 7 |
Hb_000359_320 |
0.2446954327 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 8 |
Hb_001318_290 |
0.244860078 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001227_020 |
0.2449568574 |
- |
- |
- |
| 10 |
Hb_001862_170 |
0.2601814127 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_008948_060 |
0.2611177087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001558_070 |
0.2630528086 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 13 |
Hb_005203_010 |
0.2640456088 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 14 |
Hb_031862_090 |
0.2697602242 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 15 |
Hb_001454_360 |
0.2704093687 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 16 |
Hb_002326_080 |
0.2731925603 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 17 |
Hb_001863_100 |
0.2738879414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_027445_080 |
0.2753314799 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 19 |
Hb_000165_190 |
0.2757106541 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 20 |
Hb_081448_010 |
0.2779439397 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |