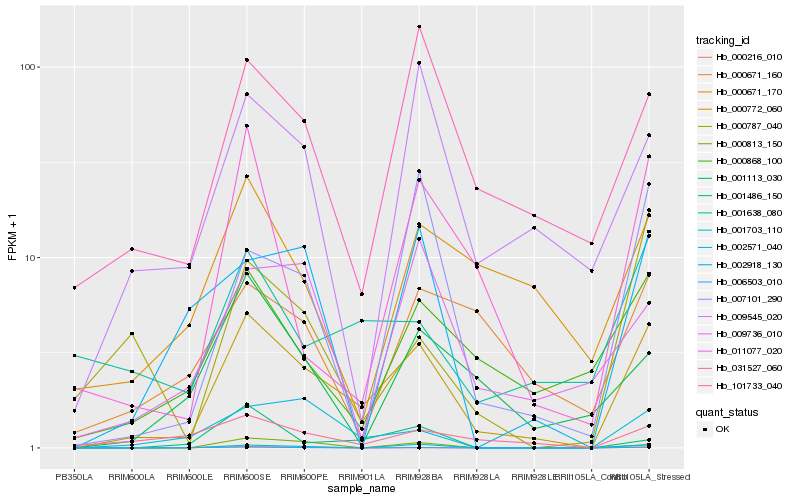
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000772_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001703_110 |
0.2525903414 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 3 |
Hb_000868_100 |
0.2664860437 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 4 |
Hb_011077_020 |
0.2665558206 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_031527_060 |
0.2683554316 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 6 |
Hb_002571_040 |
0.2691617929 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 7 |
Hb_101733_040 |
0.272781273 |
- |
- |
cytochrome P450 CYP71B63v2 [Populus trichocarpa] |
| 8 |
Hb_007101_290 |
0.2837244882 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009545_020 |
0.2880838181 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 10 |
Hb_000813_150 |
0.2897914944 |
- |
- |
- |
| 11 |
Hb_001638_080 |
0.290893525 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 12 |
Hb_002918_130 |
0.2918993654 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 13 |
Hb_001113_030 |
0.2919671359 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 14 |
Hb_009736_010 |
0.2960385774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000671_160 |
0.3032334573 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006503_010 |
0.304282658 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_000216_010 |
0.3048033308 |
- |
- |
carbon monoxide dehydrogenase [Streptomyces sp. CNQ-509] |
| 18 |
Hb_000671_170 |
0.3052861131 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_000787_040 |
0.3053520535 |
- |
- |
PREDICTED: uncharacterized protein LOC105115900 isoform X1 [Populus euphratica] |
| 20 |
Hb_001486_150 |
0.3128914275 |
- |
- |
- |