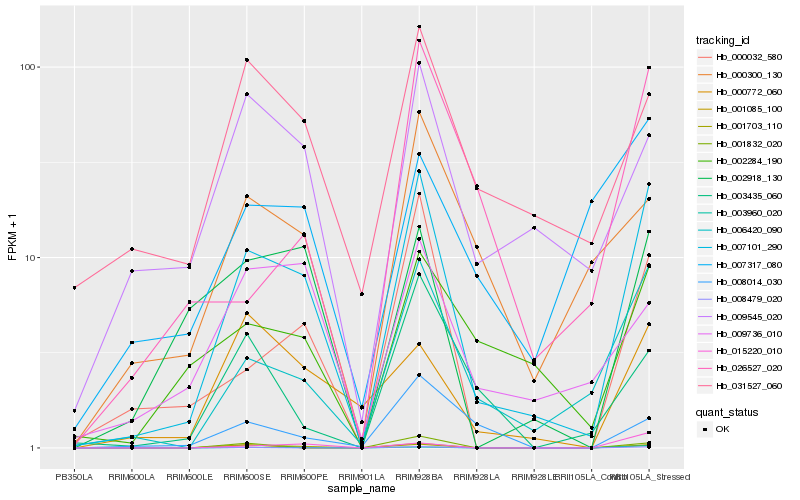
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_290 |
0.0 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001703_110 |
0.1756992155 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 3 |
Hb_006420_090 |
0.1887633123 |
- |
- |
PREDICTED: zinc finger protein 5 [Jatropha curcas] |
| 4 |
Hb_003960_020 |
0.2272986762 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000032_580 |
0.2310040228 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 6 |
Hb_002918_130 |
0.257548376 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 7 |
Hb_008479_020 |
0.2604295918 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 8 |
Hb_002284_190 |
0.262432847 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 9 |
Hb_009545_020 |
0.274428433 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 10 |
Hb_031527_060 |
0.2763457454 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 11 |
Hb_000772_060 |
0.2837244882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000300_130 |
0.2840887378 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 13 |
Hb_026527_020 |
0.2847899469 |
- |
- |
PREDICTED: putative methyltransferase DDB_G0268948 [Jatropha curcas] |
| 14 |
Hb_015220_010 |
0.2873506918 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 15 |
Hb_003435_060 |
0.288400286 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 16 |
Hb_008014_030 |
0.2905982411 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 17 |
Hb_001832_020 |
0.2949499553 |
- |
- |
PREDICTED: uncharacterized protein LOC104584029 [Brachypodium distachyon] |
| 18 |
Hb_009736_010 |
0.2953005767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001085_100 |
0.2968361706 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_007317_080 |
0.3011422966 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |