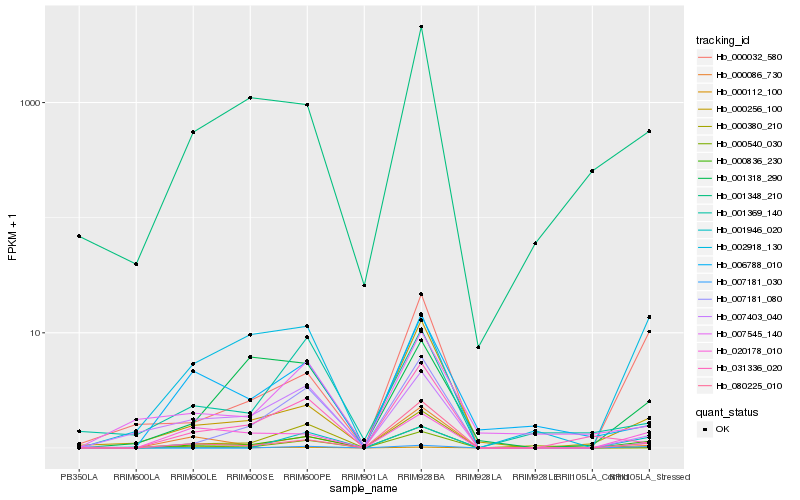
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_230 |
0.0 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 2 |
Hb_007403_040 |
0.2360239407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000032_580 |
0.2379006706 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 4 |
Hb_020178_010 |
0.2428128219 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 5 |
Hb_031336_020 |
0.2478054126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007181_080 |
0.248445538 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001348_210 |
0.2487280432 |
- |
- |
defensin protein precursor [Capsicum annuum] |
| 8 |
Hb_000086_730 |
0.2506579744 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1-like [Jatropha curcas] |
| 9 |
Hb_000380_210 |
0.2632512691 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 10 |
Hb_001946_020 |
0.2642966347 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 11 |
Hb_000112_100 |
0.2643773371 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 12 |
Hb_080225_010 |
0.264742444 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 13 |
Hb_001318_290 |
0.2652018336 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002918_130 |
0.2663617457 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 15 |
Hb_007181_030 |
0.2683537523 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000256_100 |
0.2697623238 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 17 |
Hb_001369_140 |
0.2703167561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006788_010 |
0.2721638501 |
- |
- |
PREDICTED: calcium-transporting ATPase 12, plasma membrane-type [Jatropha curcas] |
| 19 |
Hb_007545_140 |
0.2748099479 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 20 |
Hb_000540_030 |
0.2748942716 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |