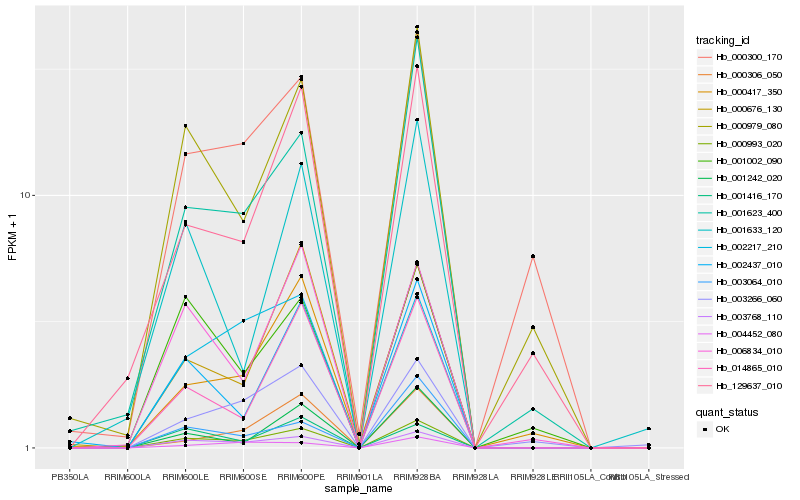
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000993_020 |
0.0 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 2 |
Hb_003768_110 |
0.0633241338 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 3 |
Hb_014865_010 |
0.1284196584 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |
| 4 |
Hb_001623_400 |
0.1294043272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002437_010 |
0.1300156652 |
- |
- |
PREDICTED: uncharacterized protein LOC105123193 [Populus euphratica] |
| 6 |
Hb_000979_080 |
0.1318799098 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 7 |
Hb_000417_350 |
0.1382607428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001242_020 |
0.1475283181 |
- |
- |
hypothetical protein JCGZ_08334 [Jatropha curcas] |
| 9 |
Hb_129637_010 |
0.151678864 |
- |
- |
PREDICTED: alpha-mannosidase isoform X1 [Populus euphratica] |
| 10 |
Hb_000676_130 |
0.1525044206 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 11 |
Hb_006834_010 |
0.1526661678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003064_010 |
0.1536577698 |
- |
- |
Senescence-related gene 1 [Theobroma cacao] |
| 13 |
Hb_002217_210 |
0.1538043795 |
- |
- |
- |
| 14 |
Hb_003266_060 |
0.1569134904 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 15 |
Hb_000306_050 |
0.1593672615 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_004452_080 |
0.1598285658 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_001416_170 |
0.1615521747 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_001002_090 |
0.1630060236 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 19 |
Hb_001633_120 |
0.1662024375 |
- |
- |
PREDICTED: uncharacterized protein LOC105630199 [Jatropha curcas] |
| 20 |
Hb_000300_170 |
0.1664609705 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |