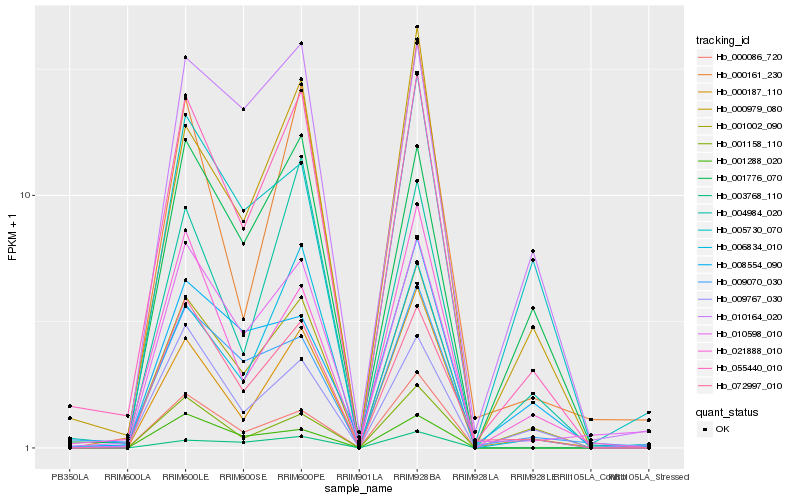
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001002_090 |
0.0 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 2 |
Hb_055440_010 |
0.0739593003 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_000979_080 |
0.1023360167 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 4 |
Hb_000187_110 |
0.1084350387 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 5 |
Hb_072997_010 |
0.1132300814 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 6 |
Hb_009767_030 |
0.1221355076 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 7 |
Hb_001776_070 |
0.1259543177 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 8 |
Hb_021888_010 |
0.1292586996 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_009070_030 |
0.1305911424 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 10 |
Hb_010164_020 |
0.1334012458 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 11 |
Hb_010598_010 |
0.1341515222 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 12 |
Hb_006834_010 |
0.1357021866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001158_110 |
0.1373893065 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 14 |
Hb_003768_110 |
0.1374811208 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 15 |
Hb_008554_090 |
0.1435932057 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000086_720 |
0.1444414506 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_005730_070 |
0.1454149351 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 18 |
Hb_004984_020 |
0.146815072 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 19 |
Hb_000161_230 |
0.1469141733 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 20 |
Hb_001288_020 |
0.1491038135 |
- |
- |
- |