| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
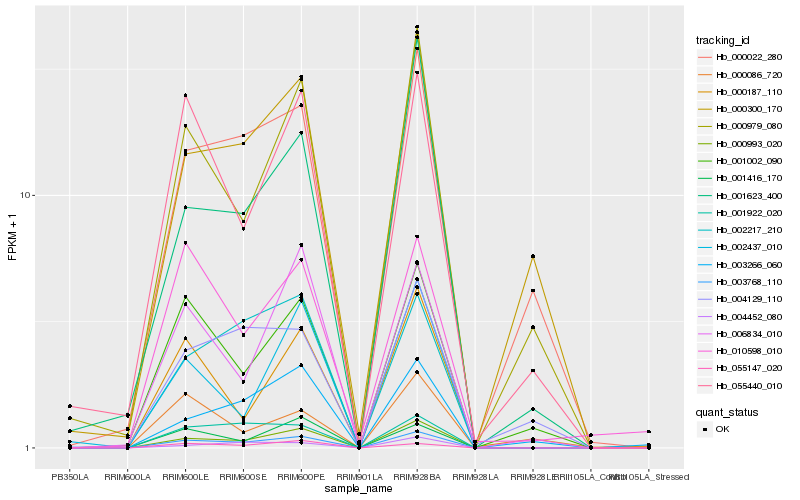
Hb_003768_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 2 |
Hb_000993_020 |
0.0633241338 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 3 |
Hb_002217_210 |
0.1283482913 |
- |
- |
- |
| 4 |
Hb_001922_020 |
0.1308201299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001002_090 |
0.1374811208 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 6 |
Hb_000979_080 |
0.1443447444 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 7 |
Hb_001416_170 |
0.1480679749 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_004452_080 |
0.1526278177 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_000022_280 |
0.1537232899 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 10 |
Hb_006834_010 |
0.1565715936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001623_400 |
0.1604484228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_055440_010 |
0.1607796732 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_055147_020 |
0.1657471796 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 14 |
Hb_002437_010 |
0.1660086616 |
- |
- |
PREDICTED: uncharacterized protein LOC105123193 [Populus euphratica] |
| 15 |
Hb_000086_720 |
0.1661925648 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 16 |
Hb_000300_170 |
0.1673507368 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 17 |
Hb_003266_060 |
0.1699228385 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 18 |
Hb_004129_110 |
0.1704594837 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 19 |
Hb_000187_110 |
0.1722187711 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 20 |
Hb_010598_010 |
0.1722508179 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |