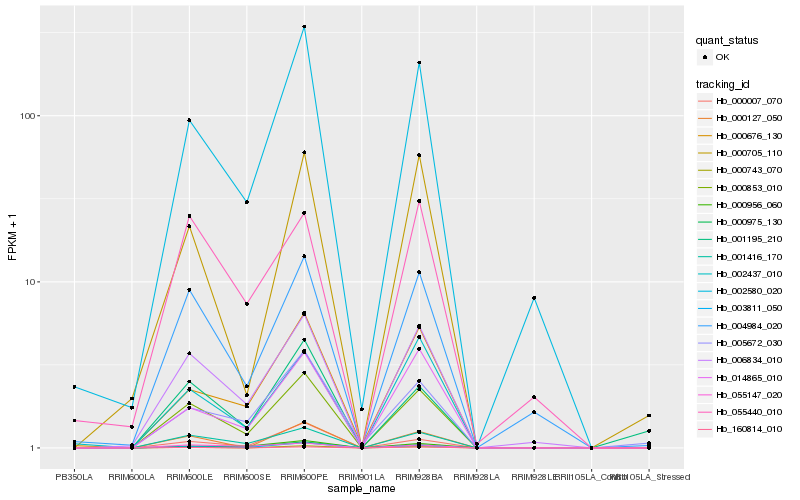
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_010 |
0.0 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 2 |
Hb_001416_170 |
0.05835253 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_006834_010 |
0.0766687563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003811_050 |
0.1152946527 |
- |
- |
hypothetical protein JCGZ_01310 [Jatropha curcas] |
| 5 |
Hb_000975_130 |
0.1153364839 |
- |
- |
PREDICTED: uncharacterized protein LOC104445138 [Eucalyptus grandis] |
| 6 |
Hb_002580_020 |
0.1185599361 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 7 |
Hb_004984_020 |
0.1202824804 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 8 |
Hb_000676_130 |
0.13445694 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 9 |
Hb_055147_020 |
0.1380801716 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 10 |
Hb_014865_010 |
0.1425072796 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |
| 11 |
Hb_005672_030 |
0.144861035 |
- |
- |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 12 |
Hb_002437_010 |
0.1469168799 |
- |
- |
PREDICTED: uncharacterized protein LOC105123193 [Populus euphratica] |
| 13 |
Hb_000956_060 |
0.153504541 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000007_070 |
0.1605512094 |
- |
- |
hypothetical protein POPTR_0008s11290g [Populus trichocarpa] |
| 15 |
Hb_000127_050 |
0.1611302448 |
- |
- |
- |
| 16 |
Hb_000705_110 |
0.1659871886 |
- |
- |
hypothetical protein POPTR_0005s09220g [Populus trichocarpa] |
| 17 |
Hb_160814_010 |
0.169847252 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_055440_010 |
0.1699358913 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_001195_210 |
0.1700861825 |
- |
- |
- |
| 20 |
Hb_000743_070 |
0.1705372484 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |