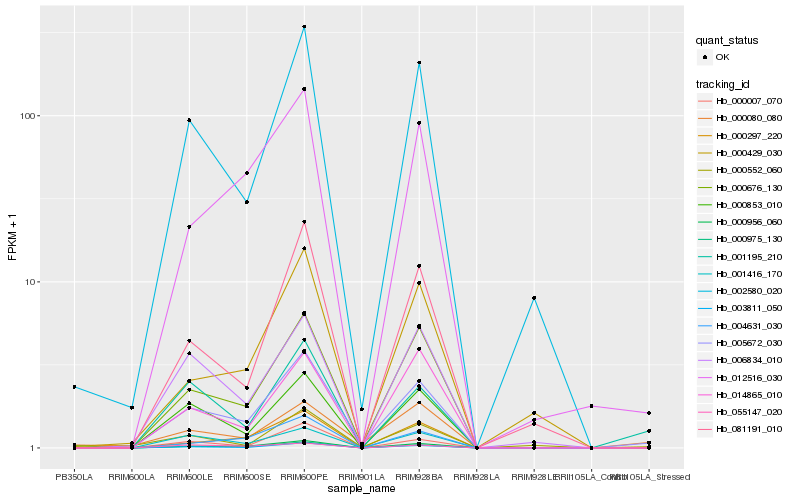
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005672_030 |
0.0 |
- |
- |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 2 |
Hb_000975_130 |
0.1099848812 |
- |
- |
PREDICTED: uncharacterized protein LOC104445138 [Eucalyptus grandis] |
| 3 |
Hb_003811_050 |
0.1146908975 |
- |
- |
hypothetical protein JCGZ_01310 [Jatropha curcas] |
| 4 |
Hb_000956_060 |
0.1223810664 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000676_130 |
0.1265143561 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 6 |
Hb_002580_020 |
0.14063094 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 7 |
Hb_000853_010 |
0.144861035 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 8 |
Hb_001195_210 |
0.1569839443 |
- |
- |
- |
| 9 |
Hb_000007_070 |
0.1606114144 |
- |
- |
hypothetical protein POPTR_0008s11290g [Populus trichocarpa] |
| 10 |
Hb_012516_030 |
0.1609252128 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 11 |
Hb_001416_170 |
0.162593017 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_081191_010 |
0.1642157231 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 13 |
Hb_006834_010 |
0.1652866989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004631_030 |
0.1664438205 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 15 |
Hb_000297_220 |
0.1674884667 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 16 |
Hb_014865_010 |
0.179147491 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |
| 17 |
Hb_055147_020 |
0.1820552245 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 18 |
Hb_000080_080 |
0.184451561 |
- |
- |
- |
| 19 |
Hb_000552_060 |
0.1853977763 |
- |
- |
PREDICTED: theobromine synthase 2-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_000429_030 |
0.1854596335 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |