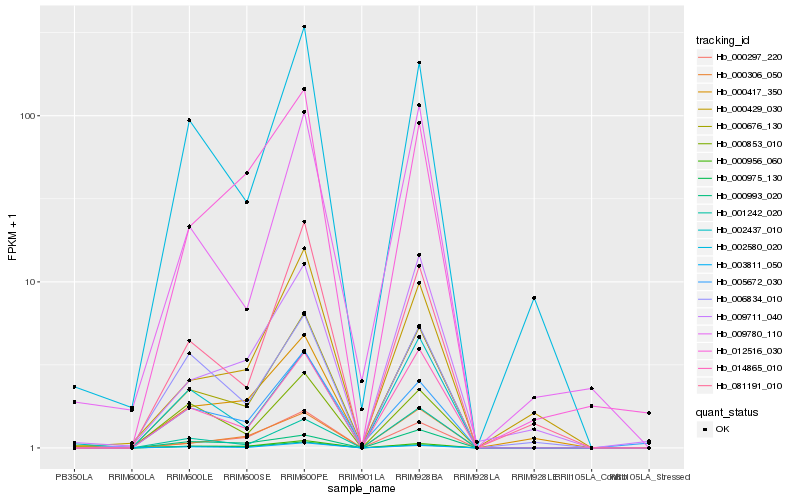
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_130 |
0.0 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 2 |
Hb_014865_010 |
0.0870623151 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |
| 3 |
Hb_000956_060 |
0.1019745957 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003811_050 |
0.1069548519 |
- |
- |
hypothetical protein JCGZ_01310 [Jatropha curcas] |
| 5 |
Hb_002580_020 |
0.1078991491 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 6 |
Hb_000975_130 |
0.1188857882 |
- |
- |
PREDICTED: uncharacterized protein LOC104445138 [Eucalyptus grandis] |
| 7 |
Hb_005672_030 |
0.1265143561 |
- |
- |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 8 |
Hb_000297_220 |
0.1272613058 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 9 |
Hb_000417_350 |
0.1294641768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002437_010 |
0.1303532062 |
- |
- |
PREDICTED: uncharacterized protein LOC105123193 [Populus euphratica] |
| 11 |
Hb_000853_010 |
0.13445694 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 12 |
Hb_081191_010 |
0.1360222536 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 13 |
Hb_009780_110 |
0.1399200023 |
- |
- |
PREDICTED: protein RSI-1 [Cicer arietinum] |
| 14 |
Hb_006834_010 |
0.1423328197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009711_040 |
0.1455910359 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 16 |
Hb_000429_030 |
0.1470203686 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000306_050 |
0.1500371402 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_012516_030 |
0.1511570994 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 19 |
Hb_000993_020 |
0.1525044206 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 20 |
Hb_001242_020 |
0.1533177107 |
- |
- |
hypothetical protein JCGZ_08334 [Jatropha curcas] |