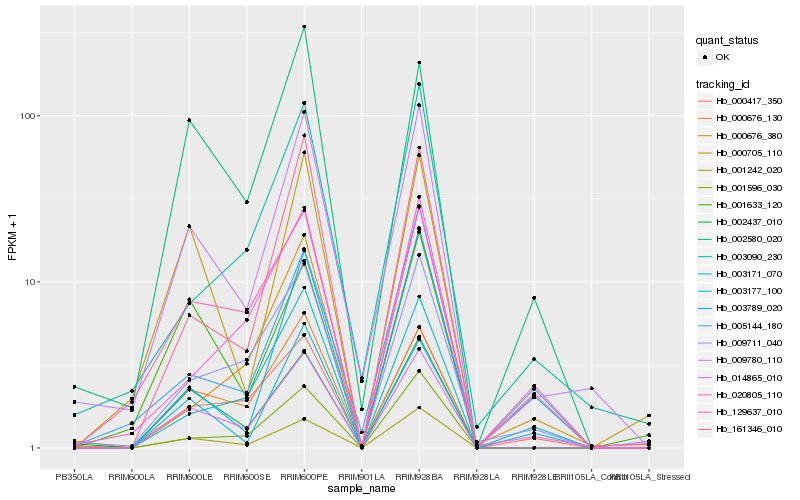
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009780_110 |
0.0 |
- |
- |
PREDICTED: protein RSI-1 [Cicer arietinum] |
| 2 |
Hb_014865_010 |
0.1225167235 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |
| 3 |
Hb_001242_020 |
0.1256746411 |
- |
- |
hypothetical protein JCGZ_08334 [Jatropha curcas] |
| 4 |
Hb_000676_130 |
0.1399200023 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 5 |
Hb_161346_010 |
0.1399328323 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 6 |
Hb_003090_230 |
0.1438442046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002437_010 |
0.1463694541 |
- |
- |
PREDICTED: uncharacterized protein LOC105123193 [Populus euphratica] |
| 8 |
Hb_001596_030 |
0.1503536085 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 9 |
Hb_000705_110 |
0.1514371558 |
- |
- |
hypothetical protein POPTR_0005s09220g [Populus trichocarpa] |
| 10 |
Hb_009711_040 |
0.1522550781 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 11 |
Hb_003177_100 |
0.1540715105 |
- |
- |
PREDICTED: auxin-induced protein 6B [Jatropha curcas] |
| 12 |
Hb_000417_350 |
0.1542829389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002580_020 |
0.1622421216 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 14 |
Hb_129637_010 |
0.1623695169 |
- |
- |
PREDICTED: alpha-mannosidase isoform X1 [Populus euphratica] |
| 15 |
Hb_003171_070 |
0.1662426483 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 16 |
Hb_005144_180 |
0.1687917263 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 17 |
Hb_020805_110 |
0.1696560874 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 18 |
Hb_000676_380 |
0.170109117 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 19 |
Hb_003789_020 |
0.1709925455 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 20 |
Hb_001633_120 |
0.1739032989 |
- |
- |
PREDICTED: uncharacterized protein LOC105630199 [Jatropha curcas] |