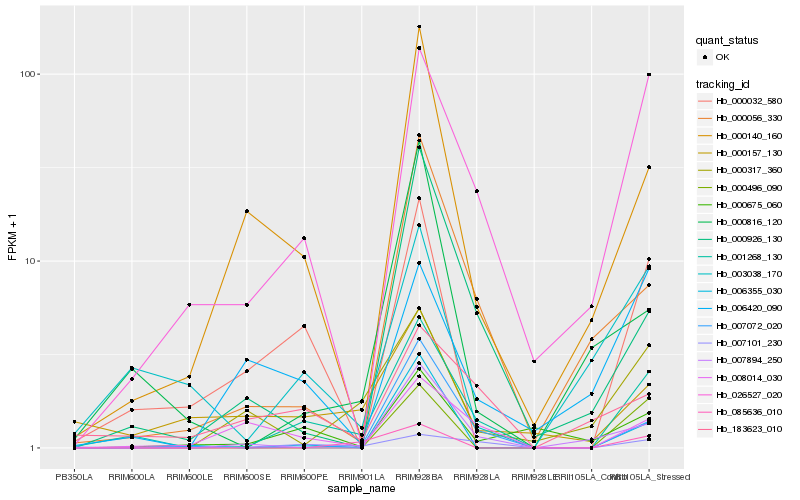
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001268_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_26837 [Jatropha curcas] |
| 2 |
Hb_026527_020 |
0.2384620966 |
- |
- |
PREDICTED: putative methyltransferase DDB_G0268948 [Jatropha curcas] |
| 3 |
Hb_007101_230 |
0.261146839 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_007072_020 |
0.2653672713 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 5 |
Hb_000032_580 |
0.2695163267 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 6 |
Hb_003038_170 |
0.2834404792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000056_330 |
0.28907338 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000140_160 |
0.2906681539 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 9 |
Hb_000926_130 |
0.2939342257 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_008014_030 |
0.2997281767 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 11 |
Hb_000816_120 |
0.3062994056 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 12 |
Hb_000675_060 |
0.3082507517 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_006355_030 |
0.3104660612 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |
| 14 |
Hb_007894_250 |
0.3107975131 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 15 |
Hb_183623_010 |
0.3144600963 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 16 |
Hb_000157_130 |
0.3179793982 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X2 [Populus euphratica] |
| 17 |
Hb_000317_360 |
0.3196499979 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g08460 [Jatropha curcas] |
| 18 |
Hb_085636_010 |
0.3224003089 |
- |
- |
PREDICTED: uncharacterized protein LOC105636643 [Jatropha curcas] |
| 19 |
Hb_006420_090 |
0.3225603896 |
- |
- |
PREDICTED: zinc finger protein 5 [Jatropha curcas] |
| 20 |
Hb_000496_090 |
0.3253733193 |
- |
- |
PREDICTED: inhibitor of trypsin and hageman factor-like [Gossypium raimondii] |