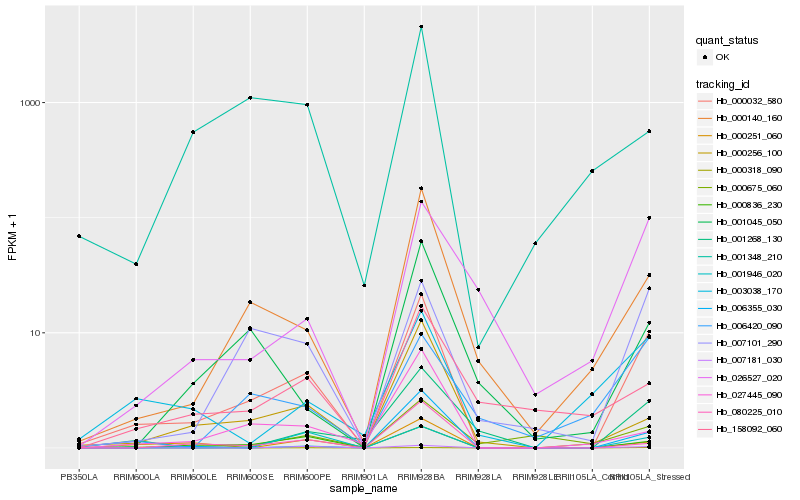
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_580 |
0.0 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 2 |
Hb_007101_290 |
0.2310040228 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000836_230 |
0.2379006706 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 4 |
Hb_000140_160 |
0.2430452015 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 5 |
Hb_001268_130 |
0.2695163267 |
- |
- |
hypothetical protein JCGZ_26837 [Jatropha curcas] |
| 6 |
Hb_000256_100 |
0.2715221267 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 7 |
Hb_026527_020 |
0.272114349 |
- |
- |
PREDICTED: putative methyltransferase DDB_G0268948 [Jatropha curcas] |
| 8 |
Hb_000251_060 |
0.275950969 |
- |
- |
PREDICTED: uncharacterized protein LOC105649757 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003038_170 |
0.2763235247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007181_030 |
0.2806569975 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 11 |
Hb_027445_090 |
0.2875949177 |
- |
- |
oleosin 18.2 kDa-like [Jatropha curcas] |
| 12 |
Hb_001045_050 |
0.289527476 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0001s33560g [Populus trichocarpa] |
| 13 |
Hb_080225_010 |
0.2899992242 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 14 |
Hb_006420_090 |
0.2904964805 |
- |
- |
PREDICTED: zinc finger protein 5 [Jatropha curcas] |
| 15 |
Hb_006355_030 |
0.2922199464 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |
| 16 |
Hb_000318_090 |
0.2976853372 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 17 |
Hb_000675_060 |
0.3018494588 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_001946_020 |
0.3037704297 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 19 |
Hb_158092_060 |
0.3074839594 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001348_210 |
0.3085695222 |
- |
- |
defensin protein precursor [Capsicum annuum] |