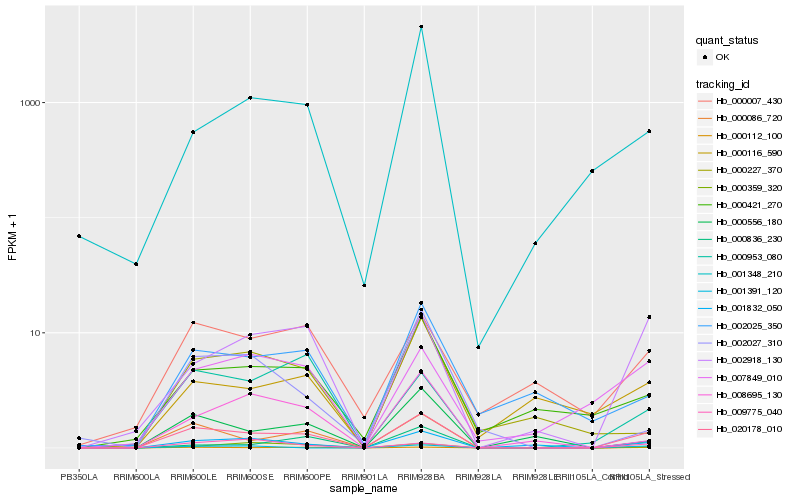
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020178_010 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 2 |
Hb_000112_100 |
0.139821847 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 3 |
Hb_001832_050 |
0.2243411425 |
- |
- |
PREDICTED: cytokinin dehydrogenase 3 [Jatropha curcas] |
| 4 |
Hb_002918_130 |
0.2267629797 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 5 |
Hb_000836_230 |
0.2428128219 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 6 |
Hb_007849_010 |
0.2487317163 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 7 |
Hb_009775_040 |
0.2556584701 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Vitis vinifera] |
| 8 |
Hb_000556_180 |
0.2561156284 |
- |
- |
hypothetical protein POPTR_0014s06690g [Populus trichocarpa] |
| 9 |
Hb_000421_270 |
0.2568745191 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 10 |
Hb_000359_320 |
0.2653552183 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 11 |
Hb_000953_080 |
0.2657201048 |
- |
- |
PREDICTED: PAN domain-containing protein At5g03700 [Jatropha curcas] |
| 12 |
Hb_002025_350 |
0.267609487 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 13 |
Hb_001348_210 |
0.2687701089 |
- |
- |
defensin protein precursor [Capsicum annuum] |
| 14 |
Hb_008695_130 |
0.2705934802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000086_720 |
0.2711406074 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 16 |
Hb_000007_430 |
0.2717384147 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 17 |
Hb_002027_310 |
0.2760263552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000116_590 |
0.2765297349 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 19 |
Hb_001391_120 |
0.2801984964 |
- |
- |
Acetyl esterase, putative [Ricinus communis] |
| 20 |
Hb_000227_370 |
0.2863263867 |
- |
- |
sugar transporter, putative [Ricinus communis] |