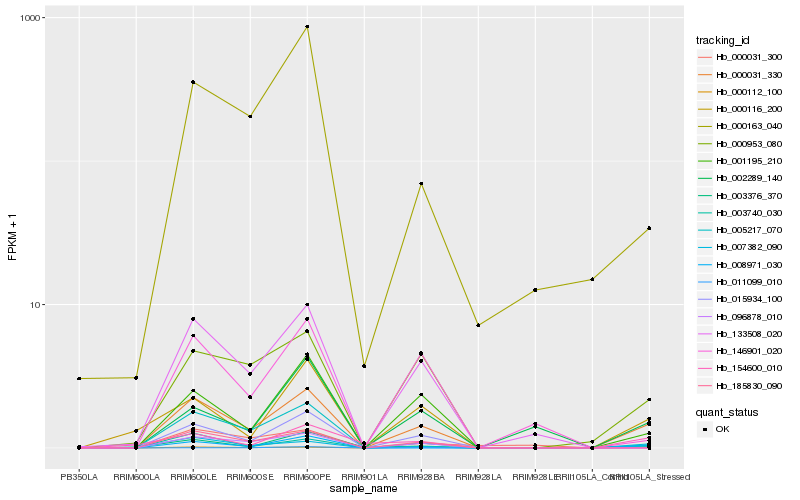
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_330 |
0.0 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0157s00200g, partial [Populus trichocarpa] |
| 2 |
Hb_007382_090 |
0.1953295501 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_001195_210 |
0.1967571232 |
- |
- |
- |
| 4 |
Hb_000953_080 |
0.1984596273 |
- |
- |
PREDICTED: PAN domain-containing protein At5g03700 [Jatropha curcas] |
| 5 |
Hb_005217_070 |
0.2007561518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003740_030 |
0.2170147528 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 7 |
Hb_000116_200 |
0.2282925662 |
transcription factor |
TF Family: HMG |
hypothetical protein POPTR_0005s10440g [Populus trichocarpa] |
| 8 |
Hb_000112_100 |
0.240999458 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 9 |
Hb_003376_370 |
0.2481700801 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_011099_010 |
0.2481871679 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_008971_030 |
0.2518325142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_185830_090 |
0.2535497835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_015934_100 |
0.2538945487 |
- |
- |
phospholipid/glycerol acyltransferase family protein [Populus trichocarpa] |
| 14 |
Hb_000163_040 |
0.2551119623 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 15 |
Hb_146901_020 |
0.2586720441 |
- |
- |
- |
| 16 |
Hb_002289_140 |
0.2587898845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_096878_010 |
0.2588810008 |
- |
- |
hypothetical protein POPTR_0112s002001g, partial [Populus trichocarpa] |
| 18 |
Hb_154600_010 |
0.2600728894 |
- |
- |
hypothetical protein JCGZ_17786 [Jatropha curcas] |
| 19 |
Hb_000031_300 |
0.2602146113 |
- |
- |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 20 |
Hb_133508_020 |
0.2618074127 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |