| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
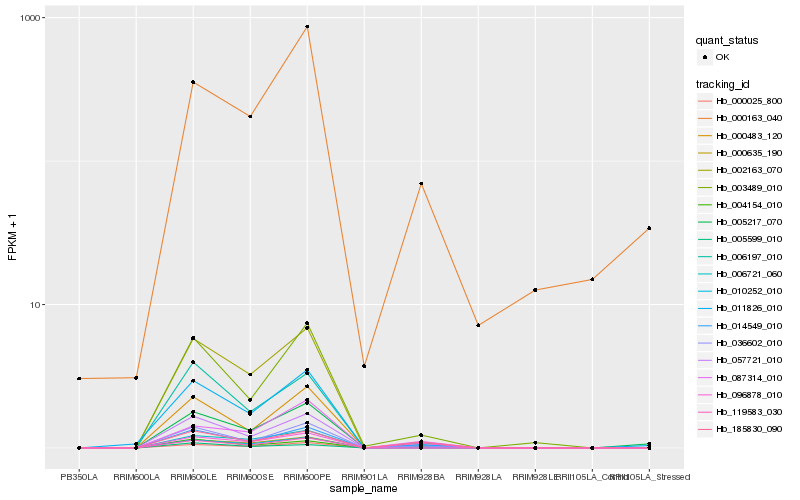
Hb_005217_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_014549_010 |
0.1299492896 |
- |
- |
PREDICTED: uncharacterized protein LOC101512634 [Cicer arietinum] |
| 3 |
Hb_011826_010 |
0.1325021564 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 4 |
Hb_036602_010 |
0.1339188553 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |
| 5 |
Hb_006721_060 |
0.1558650994 |
- |
- |
RnaseH (mitochondrion) [Malus domestica] |
| 6 |
Hb_000163_040 |
0.1563202761 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 7 |
Hb_003489_010 |
0.1625097123 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 8 |
Hb_004154_010 |
0.1628719307 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 9 |
Hb_000635_190 |
0.1682847429 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 10 |
Hb_000483_120 |
0.172600574 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0004s12530g [Populus trichocarpa] |
| 11 |
Hb_087314_010 |
0.1795259648 |
- |
- |
- |
| 12 |
Hb_096878_010 |
0.1819741288 |
- |
- |
hypothetical protein POPTR_0112s002001g, partial [Populus trichocarpa] |
| 13 |
Hb_119583_030 |
0.1822083898 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_010252_010 |
0.1835834983 |
- |
- |
- |
| 15 |
Hb_006197_010 |
0.1838793161 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 16 |
Hb_000025_800 |
0.1843466914 |
- |
- |
hypothetical protein VITISV_020143 [Vitis vinifera] |
| 17 |
Hb_002163_070 |
0.1844971266 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 18 |
Hb_185830_090 |
0.1869718721 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005599_010 |
0.1873853526 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 20 |
Hb_057721_010 |
0.1879491215 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |