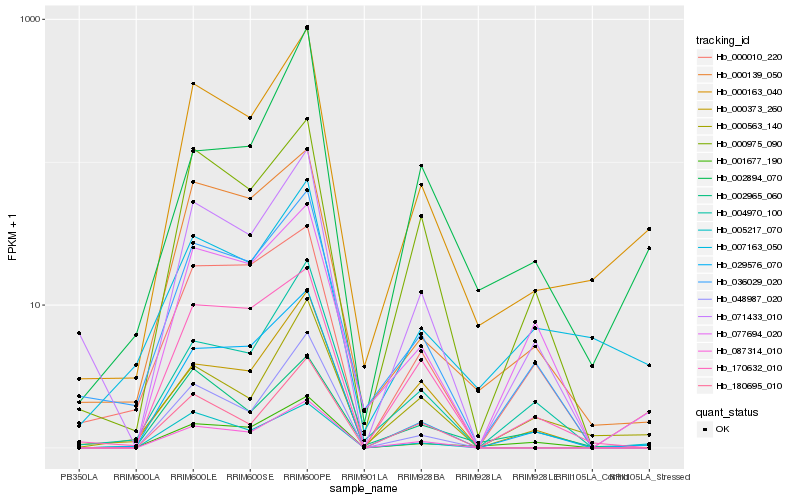
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000163_040 |
0.0 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 2 |
Hb_029576_070 |
0.1529337692 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 3 |
Hb_007163_050 |
0.1545545809 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 4 |
Hb_087314_010 |
0.1556028671 |
- |
- |
- |
| 5 |
Hb_005217_070 |
0.1563202761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000139_050 |
0.1600261953 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 7 |
Hb_000563_140 |
0.1624961906 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_036029_020 |
0.1666122935 |
- |
- |
PREDICTED: patellin-3-like [Jatropha curcas] |
| 9 |
Hb_048987_020 |
0.1720816046 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 10 |
Hb_002965_060 |
0.1725886816 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 11 |
Hb_071433_010 |
0.1726107806 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 12 |
Hb_001677_190 |
0.1746707153 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_000975_090 |
0.1808866095 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 14 |
Hb_170632_010 |
0.181783063 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_180695_010 |
0.182002157 |
- |
- |
hypothetical protein CICLE_v10013449mg [Citrus clementina] |
| 16 |
Hb_004970_100 |
0.1834682865 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 17 |
Hb_077694_020 |
0.185378161 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 18 |
Hb_002894_070 |
0.186467554 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 19 |
Hb_000373_260 |
0.1872298293 |
- |
- |
hypothetical protein POPTR_0002s06820g [Populus trichocarpa] |
| 20 |
Hb_000010_220 |
0.1875404571 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |