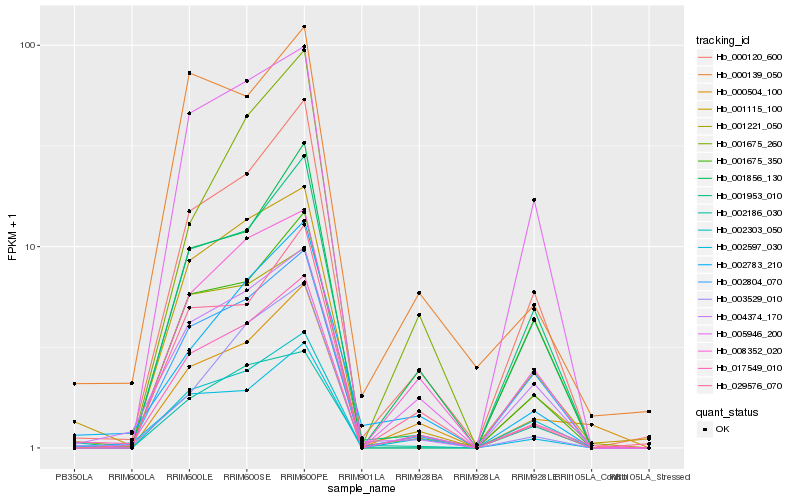
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017549_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 2 |
Hb_001115_100 |
0.0890624611 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 3 |
Hb_000120_600 |
0.095386611 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 4 |
Hb_002597_030 |
0.1024463437 |
- |
- |
- |
| 5 |
Hb_004374_170 |
0.1051848208 |
- |
- |
PREDICTED: gibberellin 20 oxidase 1-B [Jatropha curcas] |
| 6 |
Hb_002804_070 |
0.1146165124 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 7 |
Hb_029576_070 |
0.1161389302 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 8 |
Hb_003529_010 |
0.1202150031 |
- |
- |
PREDICTED: disease resistance protein RPS6-like [Jatropha curcas] |
| 9 |
Hb_001675_350 |
0.1233481907 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 10 |
Hb_002783_210 |
0.1281052423 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 11 |
Hb_008352_020 |
0.1281561952 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 12 |
Hb_001856_130 |
0.1300026918 |
- |
- |
hypothetical protein CISIN_1g022795mg [Citrus sinensis] |
| 13 |
Hb_001221_050 |
0.1373940132 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 14 |
Hb_001953_010 |
0.1390420509 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 15 |
Hb_002303_050 |
0.1391470406 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor PTL-like [Populus euphratica] |
| 16 |
Hb_000504_100 |
0.1402644876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001675_260 |
0.1407138492 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 18 |
Hb_002186_030 |
0.1414556416 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X1 [Populus euphratica] |
| 19 |
Hb_005946_200 |
0.1434726054 |
- |
- |
hypothetical protein POPTR_0006s16210g [Populus trichocarpa] |
| 20 |
Hb_000139_050 |
0.143741839 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |