| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
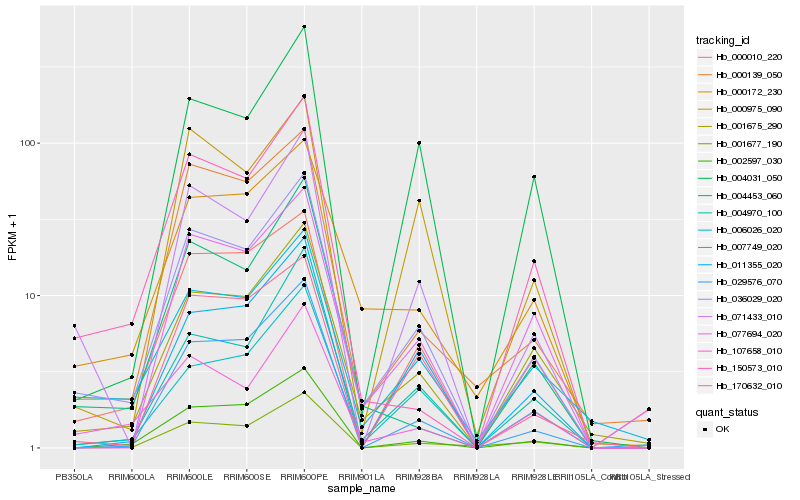
Hb_036029_020 |
0.0 |
- |
- |
PREDICTED: patellin-3-like [Jatropha curcas] |
| 2 |
Hb_071433_010 |
0.1049650012 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 3 |
Hb_001675_290 |
0.1105692623 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_007749_020 |
0.1172056414 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_000172_230 |
0.1195904443 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_002597_030 |
0.1218700298 |
- |
- |
- |
| 7 |
Hb_004970_100 |
0.1251644036 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 8 |
Hb_107658_010 |
0.1265388869 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_006026_020 |
0.1276771869 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g48800 [Jatropha curcas] |
| 10 |
Hb_000139_050 |
0.1281287888 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 11 |
Hb_004453_060 |
0.1304438601 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_000010_220 |
0.1316459467 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 13 |
Hb_001677_190 |
0.1330272636 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_029576_070 |
0.1353525792 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 15 |
Hb_011355_020 |
0.1369446302 |
- |
- |
extensin, proline-rich protein, putative [Ricinus communis] |
| 16 |
Hb_000975_090 |
0.1369464397 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 17 |
Hb_170632_010 |
0.13959817 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 18 |
Hb_004031_050 |
0.1422269513 |
- |
- |
sinapyl alcohol dehydrogenase-like protein [Populus tremula x Populus tremuloides] |
| 19 |
Hb_150573_010 |
0.1430169923 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 20 |
Hb_077694_020 |
0.1444624507 |
- |
- |
SAUR family protein [Theobroma cacao] |